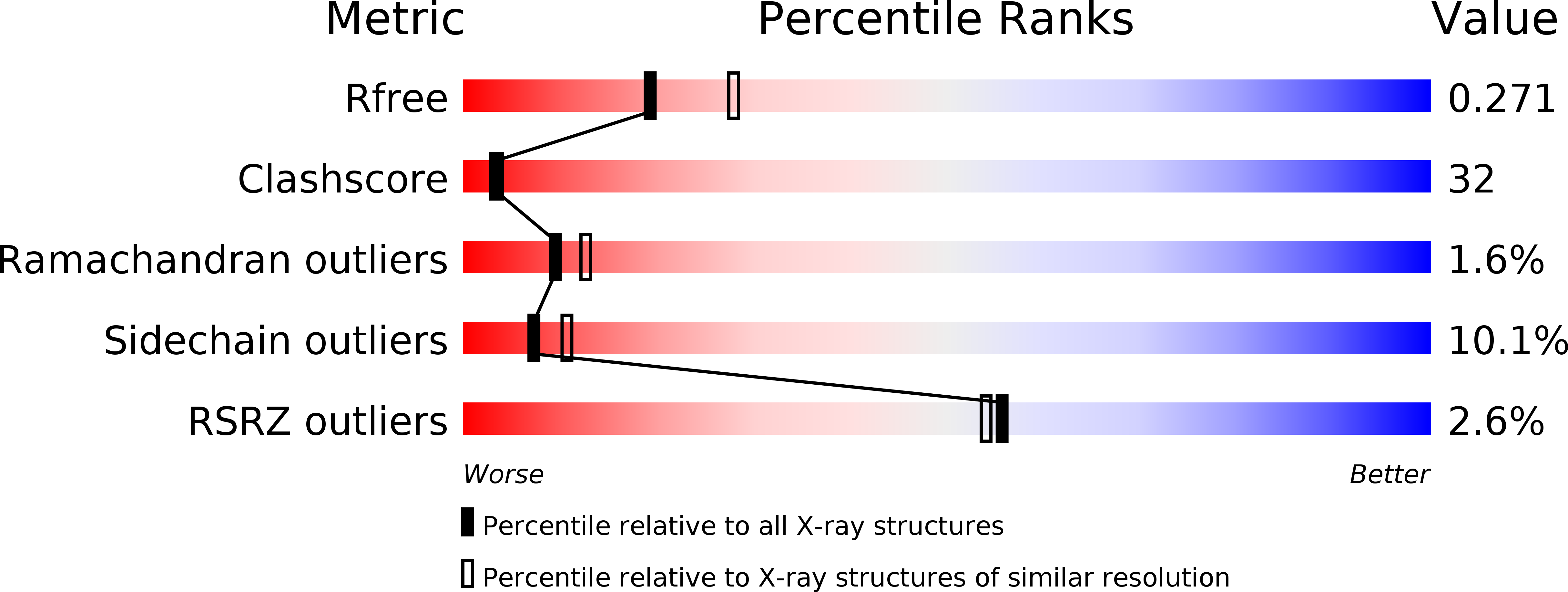
The Crystal Structure of the Hexadaca-Heme Cytochrome Hmc and a Structural Model of its Complex with Cytochrome C3
Czjzek, M., Elantak, L., Zamboni, V., Morelli, X., Dolla, A., Guerlesquin, F., Bruschi, M.(2002) Structure 10: 1677
- PubMed: 12467575
- DOI: https://doi.org/10.1016/s0969-2126(02)00909-7
- Primary Citation of Related Structures:
1GWS - PubMed Abstract:
Sulfate-reducing bacteria contain a variety of multi-heme c-type cytochromes. The cytochrome of highest molecular weight (Hmc) contains 16 heme groups and is part of a transmembrane complex involved in the sulfate respiration pathway. We present the 2.42 A resolution crystal structure of the Desulfovibrio vulgaris Hildenborough cytochrome Hmc and a structural model of the complex with its physiological electron transfer partner, cytochrome c(3), obtained by NMR restrained soft-docking calculations. The Hmc is composed of three domains, which exist independently in different sulfate-reducing species, namely cytochrome c(3), cytochrome c(7), and Hcc. The complex involves the last heme at the C-terminal region of the V-shaped Hmc and heme 4 of cytochrome c(3), and represents an example for specific cytochrome-cytochrome interaction.
Organizational Affiliation:
Architecture et Fonction des Macromolécules Biologiques, IBSM-CNRS et Université Aix-Marseille I et II, 31 Chemin Joseph-Aiguier, 13402 Marseille cedex 20, France. czjzek@afmb.cnrs-mrs.fr