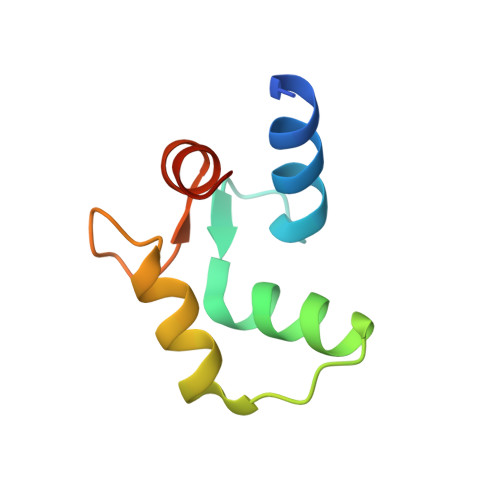
Structure of Escherichia coli fragment TR2C from calmodulin to 1.7 A resolution.
Olsson, L.L., Sjolin, L.(2001) Acta Crystallogr D Biol Crystallogr 57: 664-669
- PubMed: 11320306
- DOI: https://doi.org/10.1107/s090744490100347x
- Primary Citation of Related Structures:
1FW4 - PubMed Abstract:
Fragment TR2C is the C-terminal part of the calcium-binding protein calmodulin, including residues 78-148. The crystal structure of TR2C was solved by molecular replacement and refined to a conventional R value of 21.8% (R(free) = 22.0%), using all data in the resolution range 20.0-1.7 A. This study shows that the secondary structure of TR2C, a pair of EF-hand motifs with two calcium-binding sites, is similar to the corresponding motifs in intact calmodulin. However, it also indicates that the N-terminus of helix E is closer to the C-terminus of helix H in TR2C than in the intact protein and that the loop connecting the EF-hands shows different conformations in the two structures. The crystal structure of TR2C was further found to be similar to the set of NMR structures of this fragment, although some pronounced differences exist.
Organizational Affiliation:
Department of Inorganic Chemistry and the Center for Structural Biology, Göteborg University, SE-412 96 Göteborg, Sweden.