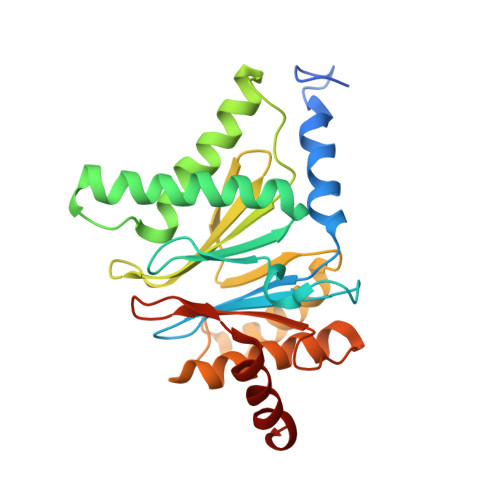
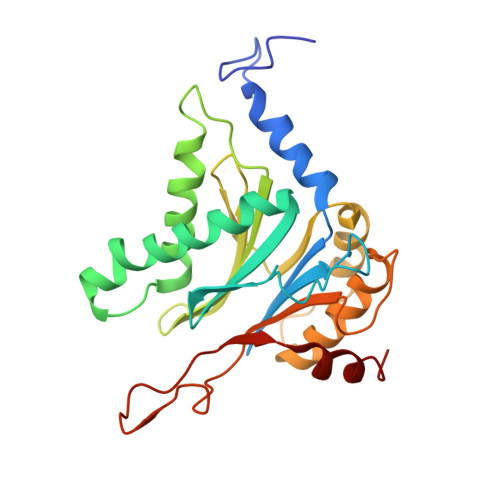
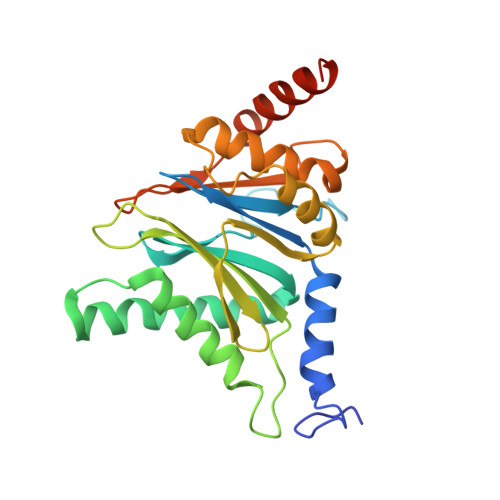
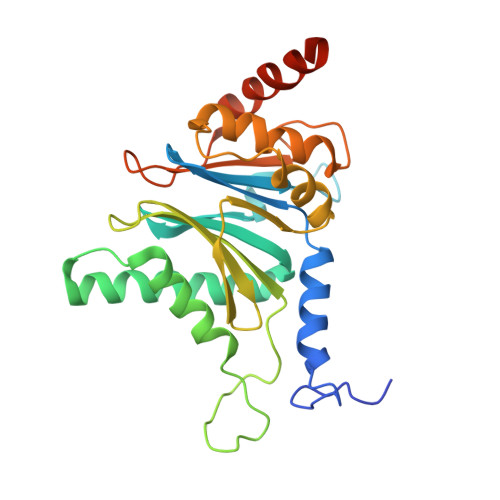
Structural basis for the activation of 20S proteasomes by 11S regulators.
Whitby, F.G., Masters, E.I., Kramer, L., Knowlton, J.R., Yao, Y., Wang, C.C., Hill, C.P.(2000) Nature 408: 115-120
- PubMed: 11081519
- DOI: https://doi.org/10.1038/35040607
- Primary Citation of Related Structures:
1FNT - PubMed Abstract:
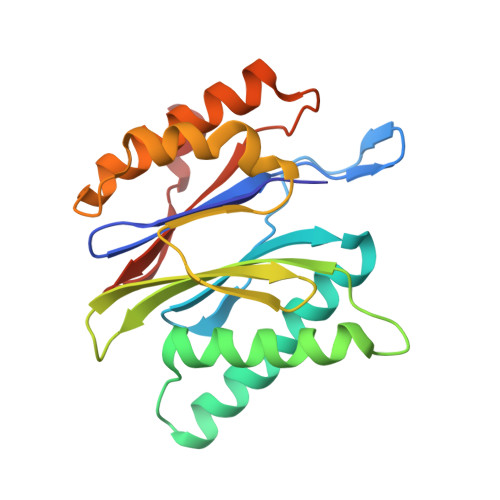
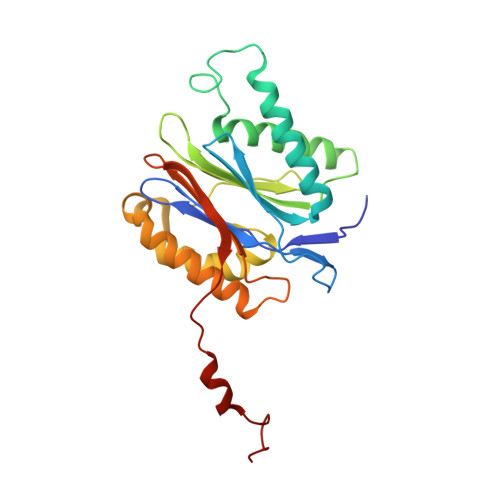
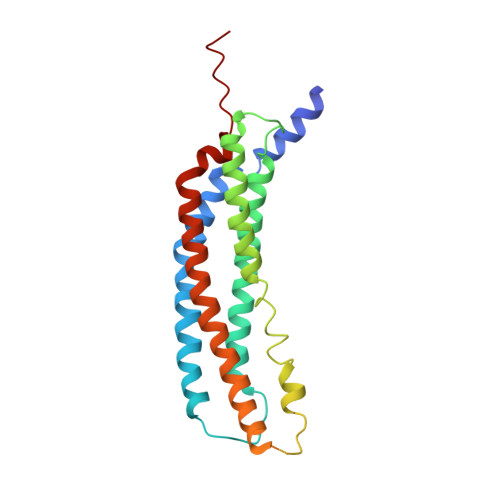
Most of the non-lysosomal proteolysis that occurs in eukaryotic cells is performed by a nonspecific and abundant barrel-shaped complex called the 20S proteasome. Substrates access the active sites, which are sequestered in an internal chamber, by traversing a narrow opening (alpha-annulus) that is blocked in the unliganded 20S proteasome by amino-terminal sequences of alpha-subunits. Peptide products probably exit the 20S proteasome through the same opening. 11S regulators (also called PA26 (ref. 4), PA28 (ref. 5) and REG) are heptamers that stimulate 20S proteasome peptidase activity in vitro and may facilitate product release in vivo. Here we report the co-crystal structure of yeast 20S proteasome with the 11S regulator from Trypanosoma brucei (PA26). PA26 carboxy-terminal tails provide binding affinity by inserting into pockets on the 20S proteasome, and PA26 activation loops induce conformational changes in alpha-subunits that open the gate separating the proteasome interior from the intracellular environment. The reduction in processivity expected for an open conformation of the exit gate may explain the role of 11S regulators in the production of ligands for major histocompatibility complex class I molecules.
- Biochemistry Department, University of Utah, Salt Lake City 84132, USA.
Organizational Affiliation: