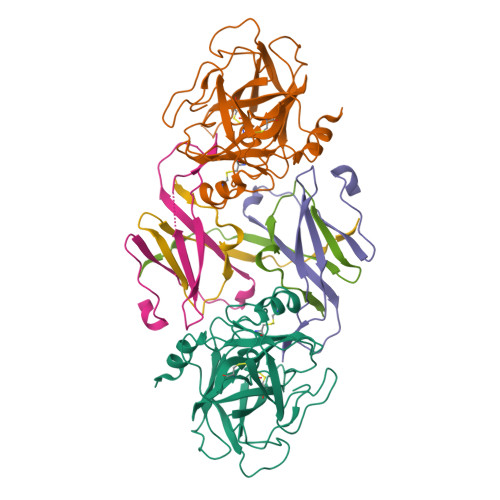
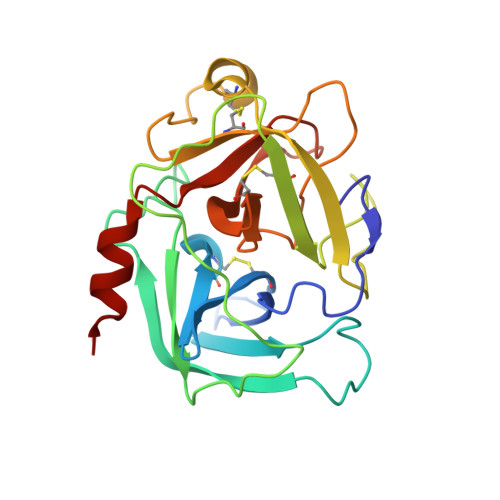
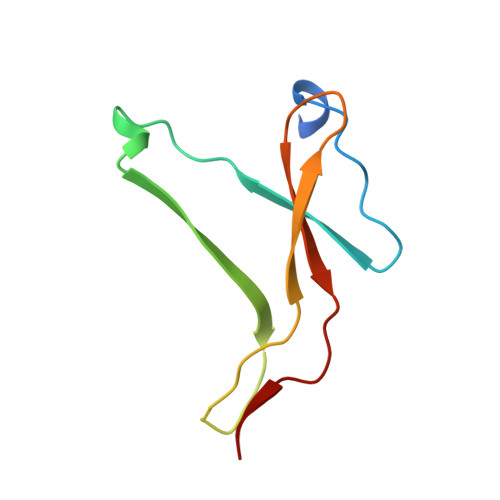
The structure of the pro-apoptotic protease granzyme B reveals the molecular determinants of its specificity
Waugh, S.M., Harris, J.L., Fletterick, R., Craik, C.S.(2000) Nat Struct Biol 7: 762-765
- PubMed: 10966646
- DOI: https://doi.org/10.1038/78992
- Primary Citation of Related Structures:
1FI8 - PubMed Abstract:
Granzyme B is a serine protease of the chymotrypsin fold that mediates cell death by cytotoxic lymphocytes. It is a processing enzyme, requiring extended peptide substrates containing an Asp residue. The determinants that allow for this substrate specificity are revealed in the three-dimensional structure of granzyme B in complex with a macromolecular inhibitor. The primary specificity for Asp occurs through a side-on interaction with Arg 226, a buried Arg side chain of granzyme B. An additional nine amino acids make contact with the substrate and define the granzyme B extended substrate specificity profile. The substrate determinants found in this structure are shared by other members of this protein class and help to reveal the properties that define substrate specificity.
Organizational Affiliation:
The Graduate Group in Biophysics, University of California, San Francisco, California 94143-0446, USA. waugh@mutant.ucsf.edu