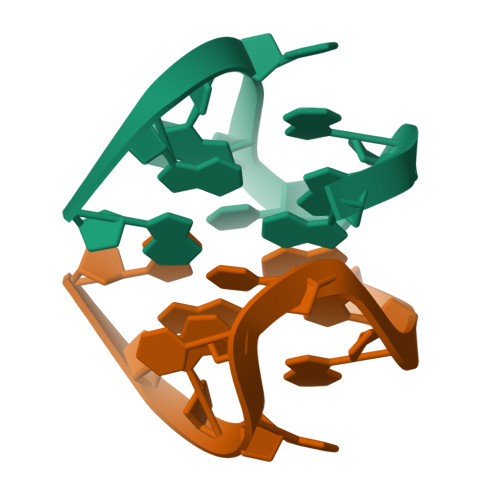
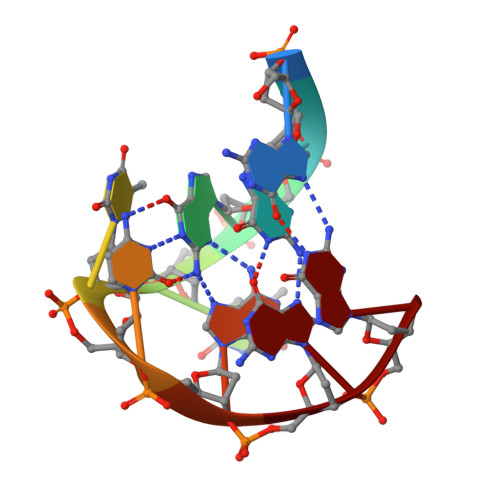
A two-stranded template-based approach to G.(C-A) triad formation: designing novel structural elements into an existing DNA framework.
Kettani, A., Basu, G., Gorin, A., Majumdar, A., Skripkin, E., Patel, D.J.(2000) J Mol Biology 301: 129-146
- PubMed: 10926497
- DOI: https://doi.org/10.1006/jmbi.2000.3932
- Primary Citation of Related Structures:
1F3S - PubMed Abstract:
We have designed a DNA sequence, d(G-G-G-T-T-C-A-G-G), which dimerizes to form a 2-fold symmetric G-quadruplex in which G(syn). G(anti).G(syn).G(anti) tetrads are sandwiched between all trans G. (C-A) triads. The NMR-based solution structural analysis was greatly aided by monitoring hydrogen bond alignments across N-H...N and N-H...O==C hydrogen bonds within the triad and tetrad, in a uniformly ((13)C,(15)N)-labeled sample of the d(G-G-G-T-T-C-A-G-G) sequence. The solution structure establishes that the guanine base-pairs with the cytosine through Watson-Crick G.C pair formation and with adenine through sheared G.A mismatch formation within the G.(C-A) triad. A model of triad DNA was constructed that contains the experimentally determined G.(C-A) triad alignment as the repeating stacked unit.
Organizational Affiliation:
Cellular Biochemistry and Biophysics Program, Memorial Sloan-Kettering Cancer Center, New York, NY 10021, USA.