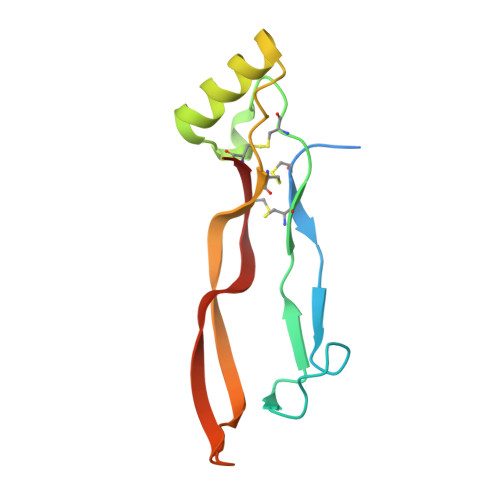
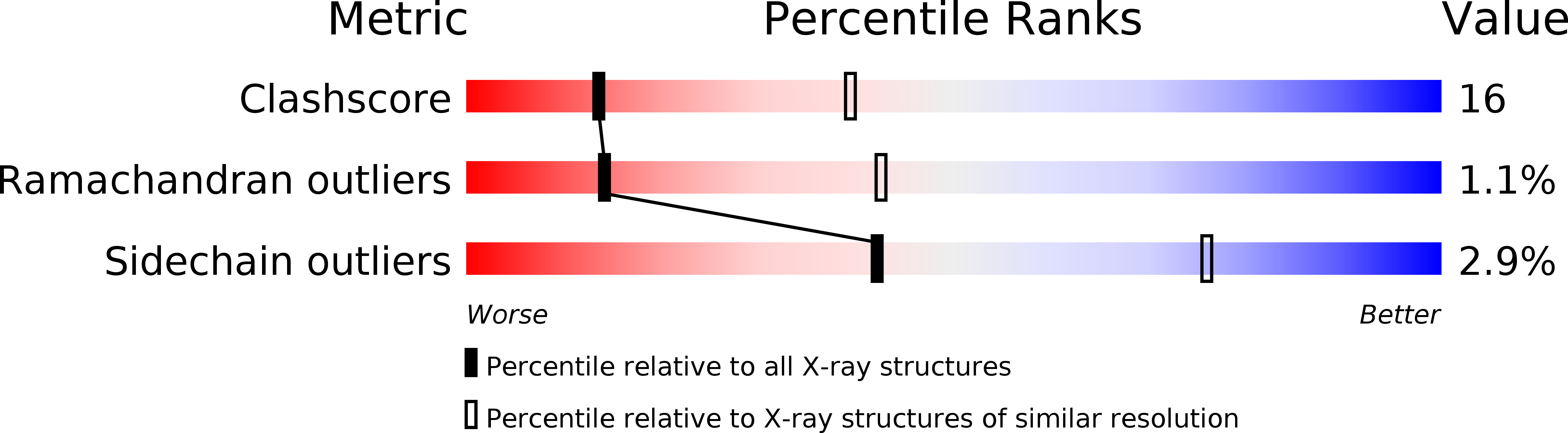Crystal structure of the BMP-2-BRIA ectodomain complex.
Kirsch, T., Sebald, W., Dreyer, M.K.(2000) Nat Struct Biol 7: 492-496
- PubMed: 10881198
- DOI: https://doi.org/10.1038/75903
- Primary Citation of Related Structures:
1ES7 - PubMed Abstract:
Bone morphogenetic proteins (BMPs) belong to the large transforming growth factor-beta (TGF-beta) superfamily of multifunctional cytokines. BMP-2 can induce ectopic bone and cartilage formation in adult vertebrates and is involved in central steps in early embryonal development in animals. Signaling by these cytokines requires binding of two types of transmembrane serine/threonine receptor kinase chains classified as type I and type II. Here we report the crystal structure of human dimeric BMP-2 in complex with two high affinity BMP receptor IA extracellular domains (BRIAec). The receptor chains bind to the 'wrist' epitopes of the BMP-2 dimer and contact both BMP-2 monomers. No contacts exist between the receptor domains. The model reveals the structural basis for discrimination between type I and type II receptors and the variability of receptor-ligand interactions that is seen in BMP-TGF-beta systems.
Organizational Affiliation:
Physiologische Chemie II, Biozentrum der Universität Würzburg, Germany.