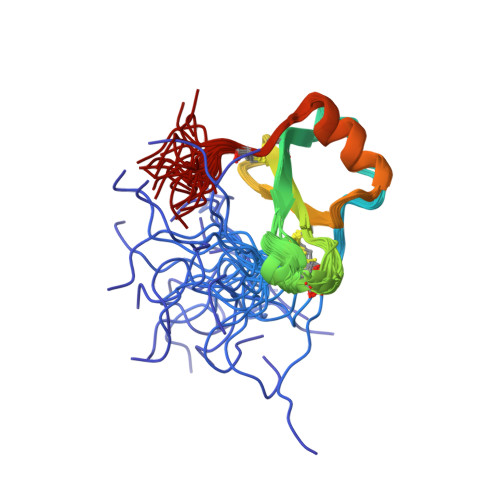
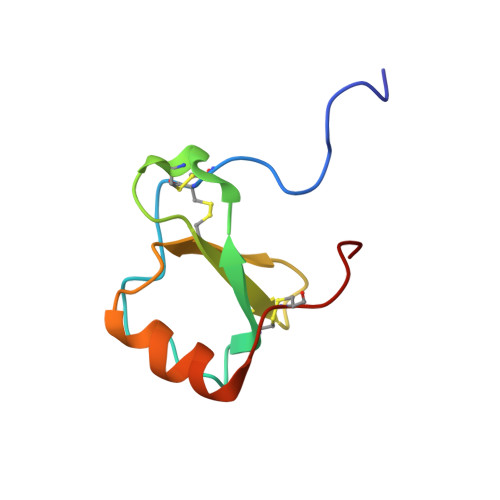
Human CC chemokine I-309, structural consequences of the additional disulfide bond.
Keizer, D.W., Crump, M.P., Lee, T.W., Slupsky, C.M., Clark-Lewis, I., Sykes, B.D.(2000) Biochemistry 39: 6053-6059
- PubMed: 10821677
- DOI: https://doi.org/10.1021/bi000089l
- Primary Citation of Related Structures:
1EL0 - PubMed Abstract:
I-309 is a member of the CC subclass of chemokines and is one of only three human chemokines known to contain an additional, third disulfide bond. The three-dimensional solution structure of I-309 was determined by (1)H nuclear magnetic resonance spectroscopy and dynamic simulated annealing. The structure of I-309, which remains monomeric at high concentrations, was determined on the basis of 978 experimental restraints. The N-terminal region of I-309 was disordered, as has been previously observed for the CC chemokine eotaxin but not others such as MCP-1 and RANTES. This was followed in I-309 by a well-ordered region between residues 13 and 69 that consisted of a 3(10)-helix, a triple-stranded antiparallel beta-sheet, and finally a C-terminal alpha-helix. Root-mean-square deviations of 0.61 and 1.16 were observed for the backbone and heavy atoms, respectively. A comparison of I-309 to eotaxin and HCC-2 revealed a significant structural change in the C-terminal region of the protein. The alpha-helix normally present in chemokines was terminated early and was followed by a short section of extended strand. These changes were a direct result of the additional disulfide bond present in this protein. An examination of the I-309 structure will aid in an understanding of the specificity of this protein with its receptor, CCR8.
Organizational Affiliation:
Protein Engineering Network Centres of Excellence (PENCE) and Department of Biochemistry, 713 Heritage Medical Research Centre, University of Alberta, Edmonton, Alberta, Canada T6G 2S2.