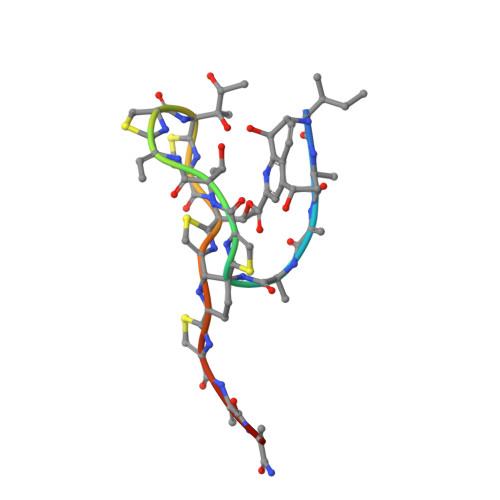
Structure of the Macrocycle Thiostrepton Solved Using the Anomalous Dispersive Contribution from Sulfur
Bond, C.S., Shaw, M.P., Alphey, M.S., Hunter, W.N.(2001) Acta Crystallogr D Biol Crystallogr 57: 755
- PubMed: 11320328
- DOI: https://doi.org/10.1107/s0907444901003134
- Primary Citation of Related Structures:
1E9W - PubMed Abstract:
The structure of a tetragonal crystal form of thiostrepton has been solved using the anomalous dispersive effects of five S atoms from high-redundancy data collected to 1.33 A resolution at the Cu Kalpha wavelength. Data measured to 1.02 A resolution with a synchrotron source were used for refinement. Details of the molecular structure, intramolecular and intermolecular interactions are given.
- The Wellcome Trust Biocentre, University of Dundee, Dundee DD1 5EH, Scotland, UK.
Organizational Affiliation: