Structure of a Dioxygen Reduction Enzyme from Desulfovibrio Gigas
Frazao, C., Silva, G., Gomes, C.M., Matias, P., Coelho, R., Sieker, L., Macedo, S., Liu, M.Y., Oliveira, S., Teixeira, M., Xavier, A.V., Rodrigues-Pousada, C., Carrondo, M.A., Le Gall, J.(2000) Nat Struct Biol 7: 1041
- PubMed: 11062560
- DOI: https://doi.org/10.1038/80961
- Primary Citation of Related Structures:
1E5D - PubMed Abstract:
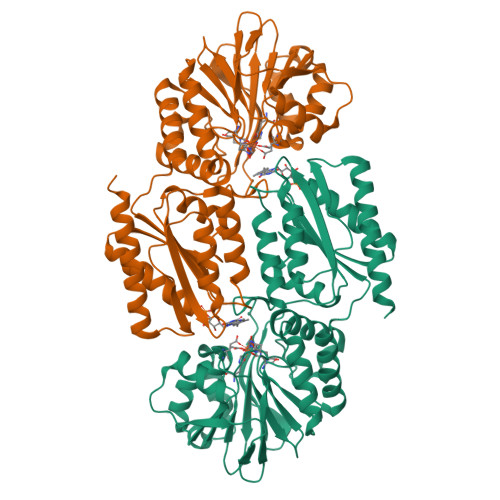
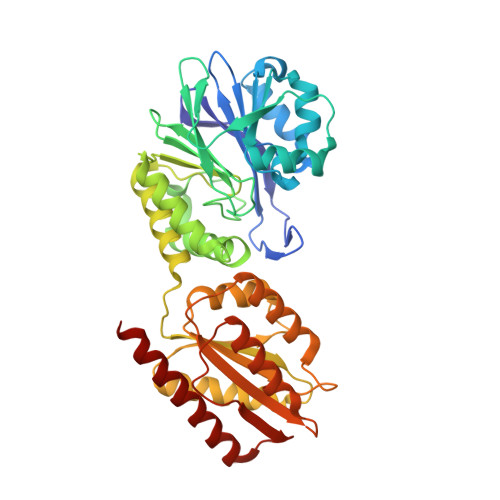
Desulfovibrio gigas is a strict anaerobe that contains a well-characterized metabolic pathway that enables it to survive transient contacts with oxygen. The terminal enzyme in this pathway, rubredoxin:oxygen oxidoreductase (ROO) reduces oxygen to water in a direct and safe way. The 2.5 A resolution crystal structure of ROO shows that each monomer of this homodimeric enzyme consists of a novel combination of two domains, a flavodoxin-like domain and a Zn-beta-lactamase-like domain that contains a di-iron center for dioxygen reduction. This is the first structure of a member of a superfamily of enzymes widespread in strict and facultative anaerobes, indicating its broad physiological significance.
Organizational Affiliation:
Instituto de Tecnologia Química e Biológica, Universidade Nova de Lisboa, Av. da República, Apartado 127, 2781-901 Oeiras, Portugal.