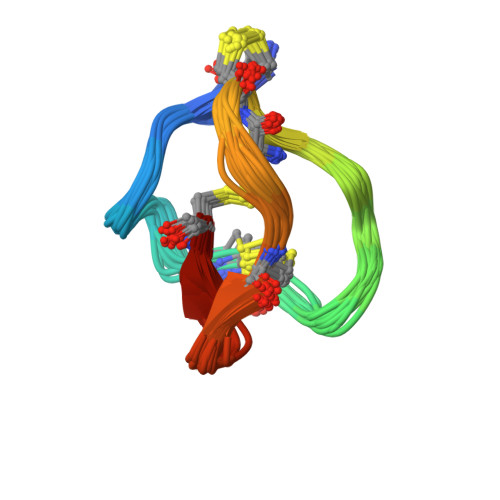
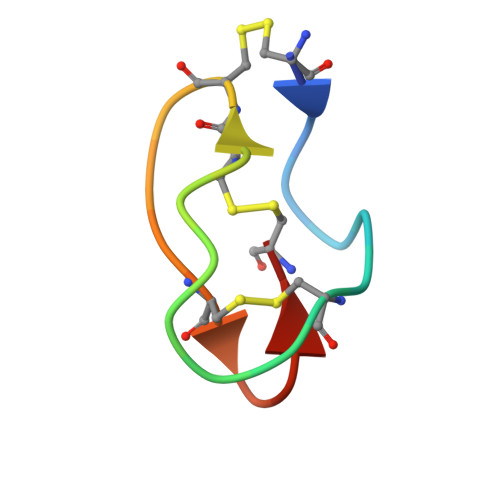
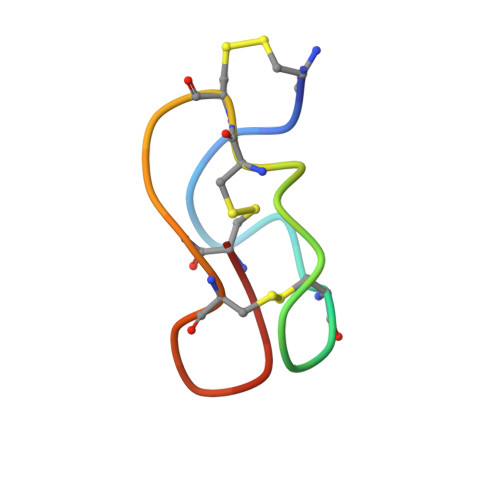
Structural and dynamic characterization of omega-conotoxin MVIIA: the binding loop exhibits slow conformational exchange.
Atkinson, R.A., Kieffer, B., Dejaegere, A., Sirockin, F., Lefevre, J.F.(2000) Biochemistry 39: 3908-3919
- PubMed: 10747778
- DOI: https://doi.org/10.1021/bi992651h
- Primary Citation of Related Structures:
1DW4, 1DW5 - PubMed Abstract:
omega-Conotoxin MVIIA is a 25-residue, disulfide-bridged polypeptide from the venom of the sea snail Conus magus that binds to neuronal N-type calcium channels. It forms a compact folded structure, presenting a loop between Cys8 and Cys15 that contains a set of residues critical for its binding. The loop does not have a unique defined structure, nor is it intrinsically flexible. Broadening of a subset of resonances in the NMR spectrum at low temperature, anomalous temperature dependence of the chemical shifts of some resonances, and exchange contributions to J(0) from (13)C relaxation measurements reveal that conformational exchange affects the residues in this loop. The effects of this exchange on the calculated structure of omega-conotoxin MVIIA are discussed. The exchange appears to be associated with a change in the conformation of the disulfide bridge Cys8-Cys20. The implications for the use of the omega-conotoxins as a scaffold for carrying other functions is discussed.
Organizational Affiliation:
UPR 9003 du CNRS, Ecole Supérieure de Biotechnologie de Strasbourg, Bld. Sébastien Brant, 67400 Illkirch, France. aatkins@nimr.mrc.ac.uk