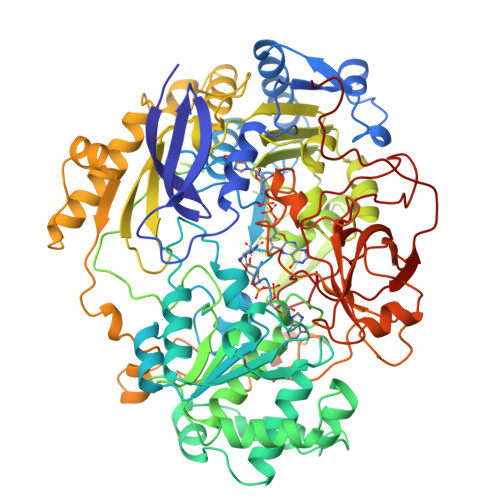
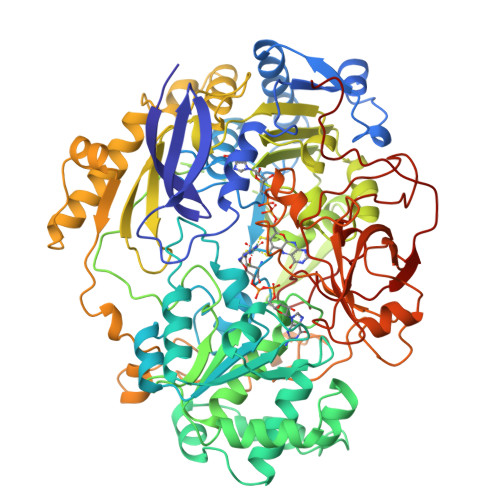
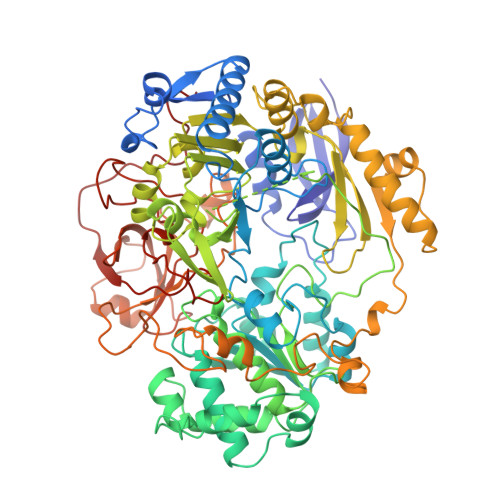
Crystal structure of dimethyl sulfoxide reductase from Rhodobacter capsulatus at 1.88 A resolution.
Schneider, F., Lowe, J., Huber, R., Schindelin, H., Kisker, C., Knablein, J.(1996) J Mol Biology 263: 53-69
- PubMed: 8890912
- DOI: https://doi.org/10.1006/jmbi.1996.0555
- Primary Citation of Related Structures:
1DMS - PubMed Abstract:
The periplasmic dimethyl sulfoxide reductase (DMSOR) from the photosynthetic purple bacterium Rhodobacter capsulatus functions as the terminal electron acceptor in its respiratory chain. The enzyme catalyzes the reduction of highly oxidized substrates like dimethyl sulfoxide to dimethyl sulfide. At a molybdenum redox center, two single electrons are transferred from cytochrome C556 to the substrate dimethyl sulfoxide, generating dimethyl sulfide and (with two protons) water. The enzyme was purified and crystallized in space group P4(1)2(1)2 with unit cell dimensions of a = b = 80.7 A and c = 229.2 A. The crystals diffract beyond 1.8 A with synchrotron radiation. The three-dimensional structure was solved by a combination of multiple isomorphous replacement and molecular replacement techniques. The atomic model was refined to an R-factor of 0.169 for 57,394 independent reflections. The spherical protein consists of four domains with a funnel-like cavity that leads to the freely accessible metal-ion redox center. The bis(molybdopterin guanine dinucleotide) molybdenum cofactor (1541 Da) of the single chain protein (85,033 Da) has the molybdenum ion bound to the cis-dithiolene group of only one molybdopterin guanine dinucleotide molecule. Three additional ligands, two oxo groups and the oxygen of a serine side-chain, are bound to the molybdenum ion. The second molybdopterin system is not part of the ligand sphere of the metal center with its sulfur atoms at distances of 3.5 A and 3.8 A away. It might be involved in electron shuttling from the protein surface to the molybdenum center.
Organizational Affiliation:
Max-Planck-Institut für Biochemie, Martinsried, FRG.