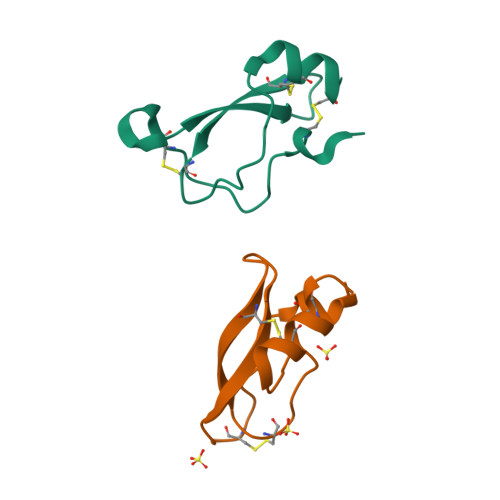
Structure of tick anticoagulant peptide at 1.6 A resolution complexed with bovine pancreatic trypsin inhibitor.
St.Charles, R., Padmanabhan, K., Arni, R.V., Padmanabhan, K.P., Tulinsky, A.(2000) Protein Sci 9: 265-272
- PubMed: 10716178
- DOI: https://doi.org/10.1110/ps.9.2.265
- Primary Citation of Related Structures:
1D0D - PubMed Abstract:
The structure of tick anticoagulant peptide (TAP) has been determined by X-ray crystallography at 1.6 A resolution complexed with bovine pancreatic trypsin inhibitor (BPTI). The TAP-BPTI crystals are tetragonal, a = b = 46.87, c = 50.35 A, space group P41, four complexes per unit cell. The TAP molecules are highly dipolar and form an intermolecular helical array along the c-axis with a diameter of about 45 A. Individual TAP units interact in a head-to-tail fashion, the positive end of one molecule associating with the distal negative end of another, and vice versa. The BPTI molecules have a uniformly distributed positively charged surface that interacts extensively through 14 hydrogen bonds and two hydrogen bonded salt bridges with the helical groove around the helical TAP chains. Comparing the structure of TAP in TAP-BPTI with TAP bound to factor Xa(Xa) suggests a massive reorganization in the N-terminal tetrapeptide and the first disulfide loop of TAP (Cys5T-Cys15T) upon binding to Xa. The Tyr1(T)OH atom of TAP moves 14.2 A to interact with Asp189 of the S1 specificity site, Arg3(T)CZ moves 5.0 A with the guanidinium group forming a cation-pi-electron complex in the S4 subsite of Xa, while Lys7(T)NZ differs in position by 10.6 A in TAP-BPTI and TAP-Xa, all of which indicates a different pre-Xa-bound conformation for the N-terminal of TAP in its native state. In contrast to TAP, the BPTI structure of TAP-BPTI is practically the same as all those of previously determined structures of BPTI, only arginine and lysine side-chain conformations showing significant differences.
Organizational Affiliation:
Department of Chemistry, Michigan State University, East Lansing 48824-1322, USA.