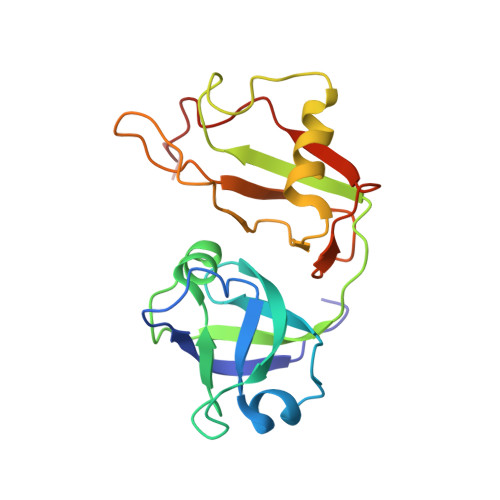
The solution structure of VAT-N reveals a 'missing link' in the evolution of complex enzymes from a simple betaalphabetabeta element.
Coles, M., Diercks, T., Liermann, J., Groger, A., Rockel, B., Baumeister, W., Koretke, K.K., Lupas, A., Peters, J., Kessler, H.(1999) Curr Biol 9: 1158-1168
- PubMed: 10531028
- DOI: https://doi.org/10.1016/S0960-9822(00)80017-2
- Primary Citation of Related Structures:
1CZ4, 1CZ5 - PubMed Abstract:
The VAT protein of the archaebacterium Thermoplasma acidophilum, like all other members of the Cdc48/p97 family of AAA ATPases, has two ATPase domains and a 185-residue amino-terminal substrate-recognition domain, VAT-N. VAT shows activity in protein folding and unfolding and thus shares the common function of these ATPases in disassembly and/or degradation of protein complexes. Using nuclear magnetic resonance (NMR) spectroscopy, we found that VAT-N is composed of two equally sized subdomains. The amino-terminal subdomain VAT-Nn (comprising residues Met1-Thr92) forms a double-psi beta-barrel whose pseudo-twofold symmetry is mirrored by an internal sequence repeat of 42 residues. The carboxy-terminal subdomain VAT-Nc (comprising residues Glu93-Gly185) forms a novel six-stranded beta-clam fold. Together, VAT-Nn and VAT-Nc form a kidney-shaped structure, in close agreement with results from electron microscopy. Sequence and structure analyses showed that VAT-Nn is related to numerous proteins including prokaryotic transcription factors, metabolic enzymes, the protease cofactors UFD1 and PrlF, and aspartic proteinases. These proteins map out an evolutionary path from simple homodimeric transcription factors containing a single copy of the VAT-Nn repeat to complex enzymes containing four copies. Our results suggest that VAT-N is a precursor of the aspartic proteinases that has acquired peptide-binding activity while remaining proteolytically incompetent. We propose that the binding site of the protein is similar to that of aspartic proteinases, in that it lies between the psi-loops of the amino-terminal beta-barrel and that it coincides with a crescent-shaped band of positive charge extending across the upper face of the molecule.
Organizational Affiliation:
Institut für Organische Chemie und Biochemie, Technische Universität München, Lichtenbergstrasse 4, 85747, Garching, Germany.