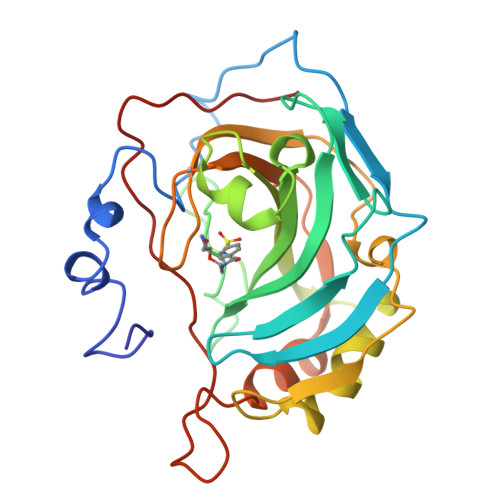
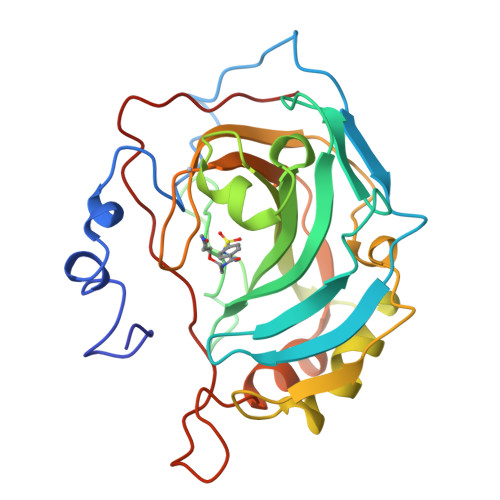
Secondary interactions significantly removed from the sulfonamide binding pocket of carbonic anhydrase II influence inhibitor binding constants.
Boriack, P.A., Christianson, D.W., Kingery-Wood, J., Whitesides, G.M.(1995) J Med Chem 38: 2286-2291
- PubMed: 7608893
- DOI: https://doi.org/10.1021/jm00013a004
- Primary Citation of Related Structures:
1CNW, 1CNX, 1CNY - PubMed Abstract:
A series of competitive inhibitors of carbonic anhydrase II (CAII; EC 4.2.1.1) that consists of oligo(ethylene glycol) units attached to p-benzenesulfonamides with pendant amino acids, H2NSO2C6H4CONHCH2CH2OCH2CH2OCH2CH2NHCOCHRNH3+, have been synthesized and examined using competitive fluorescence assays. Three of the strongest inhibitors, designated EG3NH3+, EG3GlyNH3+, and EG3PheNH3+, have been studied by X-ray crystallographic methods at limiting resolutions of 1.9, 2.0, and 2.3 A, respectively. The sulfonamide-zinc binding modes and the association of the ethylene glycol linkers to the hydrophobic patch of the active site are similar in all three inhibitors. Differences in the values of Kd are therefore not due to differences in zinc coordination or to differences in the modes of enzyme-glycol association but instead appear to arise from interaction of the pendant amino acids with the surface of the protein. These pendant groups are, however, not sufficiently ordered to be visible in electron density maps. Thus, structural variations of inhibitors at locations distant from the primary binding (i.e., the sulfonamide group) site affect the overall binding affinities of inhibitors (e.g., Kd (EG3PheNH3+) = 14 nM as compared with Kd (EG3GluNH3+) = 100 nM).
Organizational Affiliation:
Department of Chemistry, University of Pennsylvania, Philadelphia 19104-6323, USA.