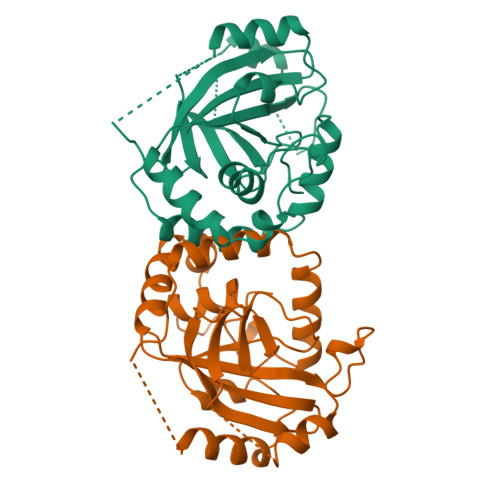
Three-dimensional structure of human cytomegalovirus protease.
Shieh, H.S., Kurumbail, R.G., Stevens, A.M., Stegeman, R.A., Sturman, E.J., Pak, J.Y., Wittwer, A.J., Palmier, M.O., Wiegand, R.C., Holwerda, B.C., Stallings, W.C.(1996) Nature 383: 279-282
- PubMed: 8805708
- DOI: https://doi.org/10.1038/383279a0
- Primary Citation of Related Structures:
1CMV - PubMed Abstract:
Herpesviruses encode a serine protease that specifically cleaves assembly protein. This protease is critical for replication, and represents a new target for antiviral drug design. Here we report the three-dimensional structure of the protease from human cytomegalovirus (hCMV) at 2.27 angstroms resolution. The structure reveals a unique fold and new catalytic strategy for cleavage. The monomer fold of the enzyme, a seven-stranded beta-barrel encircled by a chain of helices that form the carboxy terminus of the molecule, is unrelated to those observed in classic serine proteases such as chymotrypsin and subtilisin. The serine nucleophile at position 132 is activated by two juxtaposed histidine residues at positions 63 and 157. Dimerization, which seems to be necessary for activity, is observed in the crystals. Correlations of the structure with the sequences of herpesvirus proteases suggest that dimerization may confer specificity and recognition in substrate binding.
Organizational Affiliation:
Department of Medicinal and Structural Chemistry, Monsanto/Searle, Searle Discovery Research, St Louis, Missouri 63198, USA. hsshie@ccmail.monsanto.com