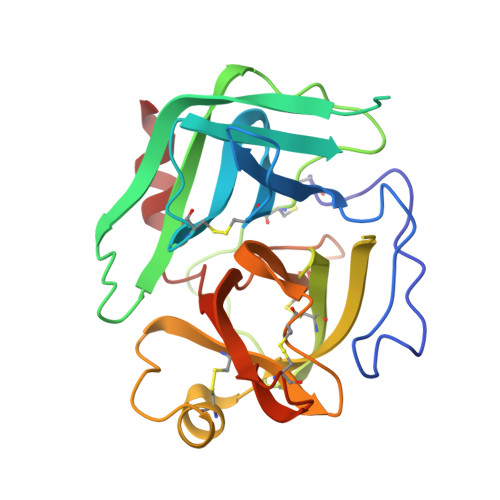
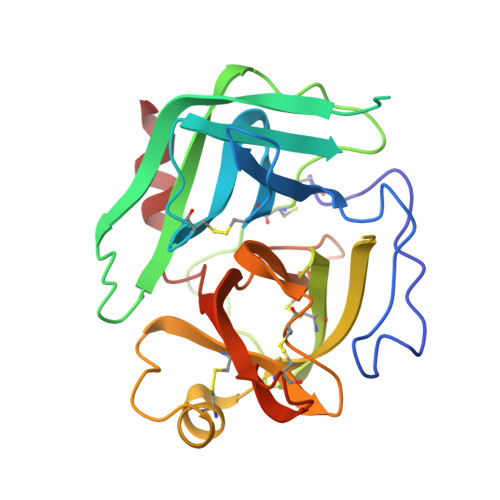
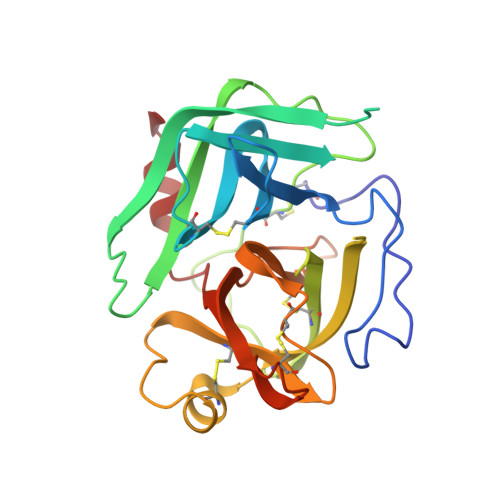
Chymotrypsinogen: 2.5-angstrom crystal structure, comparison with alpha-chymotrypsin, and implications for zymogen activation.
Freer, S.T., Kraut, J., Robertus, J.D., Wright, H.T., Xuong, N.H.(1970) Biochemistry 9: 1997-2009
- PubMed: 5442169
- DOI: https://doi.org/10.1021/bi00811a022
- Primary Citation of Related Structures:
1CHG