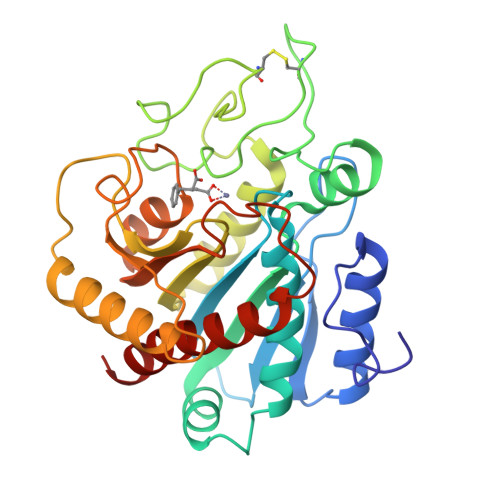
Crystal structure of the complex between carboxypeptidase A and the biproduct analog inhibitor L-benzylsuccinate at 2.0 A resolution.
Mangani, S., Carloni, P., Orioli, P.(1992) J Mol Biology 223: 573-578
- PubMed: 1738164
- DOI: https://doi.org/10.1016/0022-2836(92)90671-6
- Primary Citation of Related Structures:
1CBX - PubMed Abstract:
The X-ray crystal structure of the carboxypeptidase A-L-benzylsuccinate complex has been refined at 2.0 A resolution to a final R-factor of 0.166. One molecule of the inhibitor binds to the enzyme active site. The terminal carboxylate forms a salt link with the guanidinium group of Arg145 and hydrogen bonds with Tyr248 and Asn144. The second carboxylate group binds to the zinc ion in an asymmetric bidentate fashion replacing the water molecule of the native structure. The zinc ion moves 0.5 A from its position in the native structure to accommodate the inhibitor binding. The overall stereochemistry around the zinc can be considered a distorted tetrahedron, although six atoms of the co-ordinated groups lie within 3.0 A from the zinc ion. The key for the strong inhibitory properties of L-benzylsuccinate can be found in its ability both to co-ordinate the zinc and to form a short carboxyl-carboxylate-type hydrogen bond (2.5 A) with Glu270.
Organizational Affiliation:
Dipartimento di Chimica, Università di Siena, Italy.