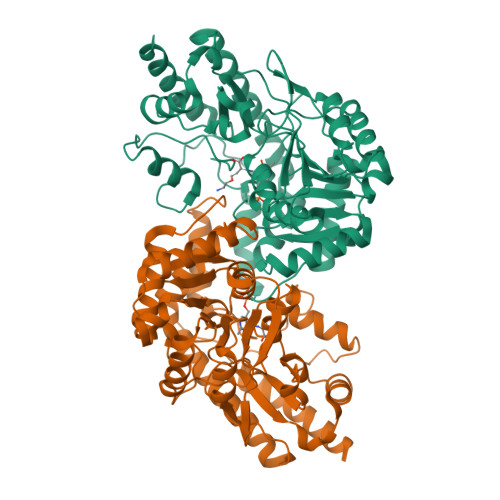
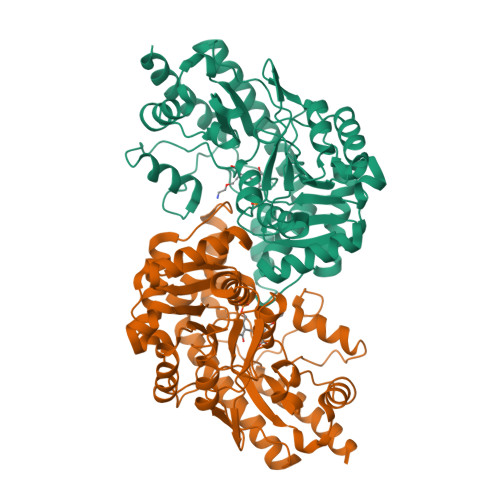
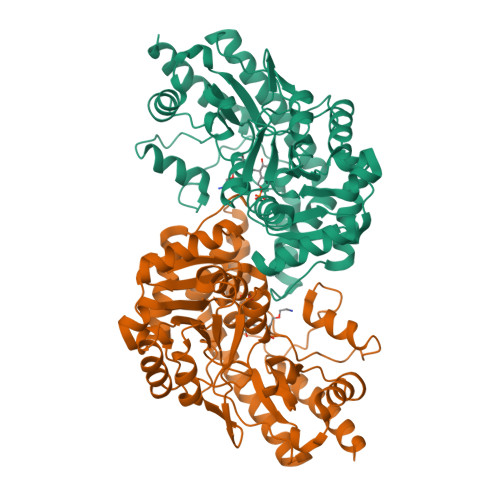
Crystal structure of cystalysin from Treponema denticola: a pyridoxal 5'-phosphate-dependent protein acting as a haemolytic enzyme.
Krupka, H.I., Huber, R., Holt, S.C., Clausen, T.(2000) EMBO J 19: 3168-3178
- PubMed: 10880431
- DOI: https://doi.org/10.1093/emboj/19.13.3168
- Primary Citation of Related Structures:
1C7N, 1C7O - PubMed Abstract:
Cystalysin is a C(beta)-S(gamma) lyase from the oral pathogen Treponema denticola catabolyzing L-cysteine to produce pyruvate, ammonia and H(2)S. With its ability to induce cell lysis, cystalysin represents a new class of pyridoxal 5'-phosphate (PLP)-dependent virulence factors. The crystal structure of cystalysin was solved at 1.9 A resolution and revealed a folding and quaternary arrangement similar to aminotransferases. Based on the active site architecture, a detailed catalytic mechanism is proposed for the catabolism of S-containing amino acid substrates yielding H(2)S and cysteine persulfide. Since no homologies were observed with known haemolysins the cytotoxicity of cystalysin is attributed to this chemical reaction. Analysis of the cystalysin-L-aminoethoxyvinylglycine (AVG) complex revealed a 'dead end' ketimine PLP derivative, resulting in a total loss of enzyme activity. Cystalysin represents an essential factor of adult periodontitis, therefore the structure of the cystalysin-AVG complex may provide the chemical basis for rational drug design.
Organizational Affiliation:
Max-Planck-Institut für Biochemie, Abteilung Strukturforschung, Am Klopferspitz 18a, 82152 Martinsried, Germany.