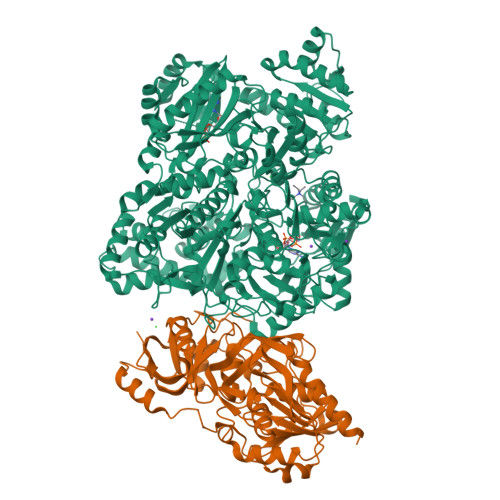
The small subunit of carbamoyl phosphate synthetase: snapshots along the reaction pathway.
Thoden, J.B., Huang, X., Raushel, F.M., Holden, H.M.(1999) Biochemistry 38: 16158-16166
- PubMed: 10587438
- DOI: https://doi.org/10.1021/bi991741j
- Primary Citation of Related Structures:
1C30, 1C3O, 1CS0 - PubMed Abstract:
Carbamoyl phosphate synthetase (CPS) plays a key role in both arginine and pyrimidine biosynthesis by catalyzing the production of carbamoyl phosphate. The enzyme from Escherichi coli consists of two polypeptide chains referred to as the small and large subunits. On the basis of both amino acid sequence analyses and X-ray structural studies, it is known that the small subunit belongs to the Triad or Type I class of amidotransferases, all of which contain a cysteine-histidine (Cys269 and His353) couple required for activity. The hydrolysis of glutamine by the small subunit has been proposed to occur via two tetrahedral intermediates and a glutamyl-thioester moiety. Here, we describe the three-dimensional structures of the C269S/glutamine and CPS/glutamate gamma-semialdehyde complexes, which serve as mimics for the Michaelis complex and the tetrahedral intermediates, respectively. In conjunction with the previously solved glutamyl-thioester intermediate complex, the stereochemical course of glutamine hydrolysis in CPS has been outlined. Specifically, attack by the thiolate of Cys269 occurs at the Si face of the carboxamide group of the glutamine substrate leading to a tetrahedral intermediate with an S-configuration. Both the backbone amide groups of Gly241 and Leu270, and O(gamma) of Ser47 play key roles in stabilizing the developing oxyanion. Collapse of the tetrahedral intermediate leads to formation of the glutamyl-thioester intermediate, which is subsequently attacked at the Si face by an activated water molecule positioned near His353. The results described here serve as a paradigm for other members of the Triad class of amidotranferases.
Organizational Affiliation:
Department of Biochemistry, University of Wisconsin-Madison 53705, USA.