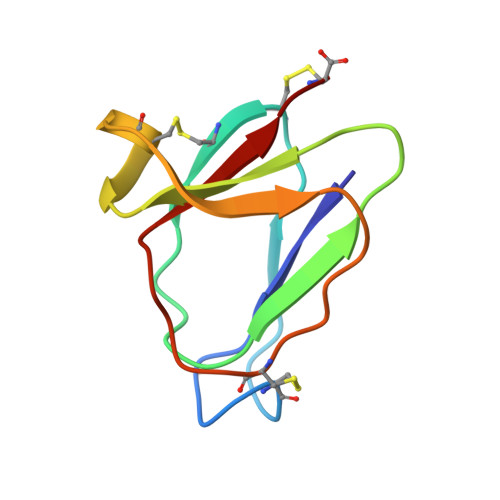
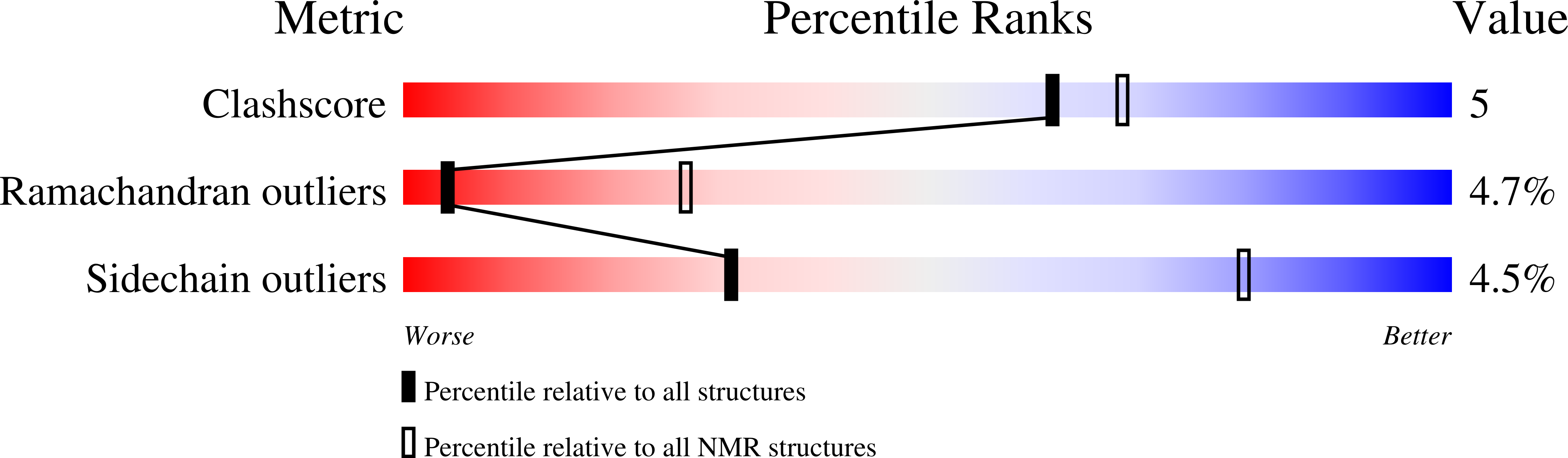MiAMP1, a novel protein from Macadamia integrifolia adopts a Greek key beta-barrel fold unique amongst plant antimicrobial proteins.
McManus, A.M., Nielsen, K.J., Marcus, J.P., Harrison, S.J., Green, J.L., Manners, J.M., Craik, D.J.(1999) J Mol Biol 293: 629-638
- PubMed: 10543955
- DOI: https://doi.org/10.1006/jmbi.1999.3163
- Primary Citation of Related Structures:
1C01 - PubMed Abstract:
MiAMP1 is a recently discovered 76 amino acid residue, highly basic protein from the nut kernel of Macadamia integrifolia which possesses no sequence homology to any known protein and inhibits the growth of several microbial plant pathogens in vitro while having no effect on mammalian or plant cells. It is considered to be a potentially useful tool for the genetic engineering of disease resistance in transgenic crop plants and for the design of new fungicides. The three-dimensional structure of MiAMP1 was determined through homonuclear and heteronuclear ((15)N) 2D NMR spectroscopy and subsequent simulated annealing calculations with the ultimate aim of understanding the structure-activity relationships of the protein. MiAMP1 is made up of eight beta-strands which are arranged in two Greek key motifs. These Greek key motifs associate to form a Greek key beta-barrel. This structure is unique amongst plant antimicrobial proteins and forms a new class which we term the beta-barrelins. Interestingly, the structure of MiAMP1 bears remarkable similarity to a yeast killer toxin from Williopsis mrakii. This toxin acts by inhibiting beta-glucan synthesis and thereby cell wall construction in sensitive strains of yeast. The structural similarity of MiAMP1 and WmKT, which originate from plant and fungal phyla respectively, may reflect a similar mode of action.
Organizational Affiliation:
Centre for Drug Design and Development, The University of Queensland, Brisbane, Queensland, 4072, Australia.