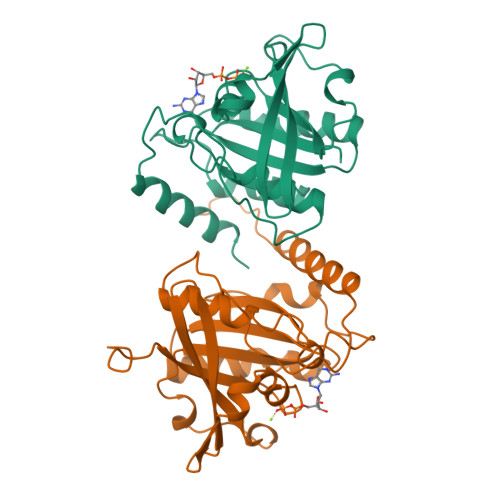
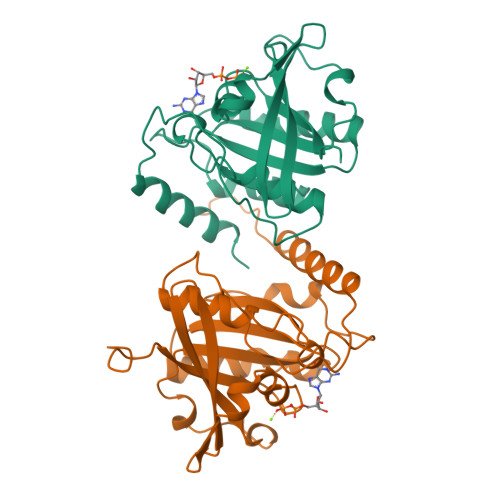
The structure of the Q69L mutant of GDP-Ran shows a major conformational change in the switch II loop that accounts for its failure to bind nuclear transport factor 2 (NTF2).
Stewart, M., Kent, H.M., McCoy, A.J.(1998) J Mol Biology 284: 1517-1527
- PubMed: 9878368
- DOI: https://doi.org/10.1006/jmbi.1998.2204
- Primary Citation of Related Structures:
1BYU, 3RAN - PubMed Abstract:
We report the 2.3 A resolution X-ray crystal structure of the GDP-bound form of the RanQ69L mutant that is used extensively in studies of nucleocytoplasmic transport and cell-cycle progression. When the structure of GDP-RanQ69L from monoclinic crystals with P21 symmetry was compared with the structure of wild-type Ran obtained from monoclinic crystals, the Q69L mutant showed a large conformational change in residues 68-74, which are in the switch II region of the molecule which changes conformation in response to nucleotide state and which forms the major interaction interface with nuclear transport factor 2 (NTF2, sometimes called p10). This conformational change alters the positions of key residues such as Lys71, Phe72 and Arg76 that are crucial for the interaction of GDP-Ran with NTF2 and indeed, solution binding studies were unable to detect any interaction between NTF2 and GDP-RanQ69L under conditions where GDP-Ran bound effectively. This interaction between NTF2 and GDP-Ran is required for efficient nuclear protein import and may function between the docking and translocation steps of the pathway.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Hills Rd., Cambridge, CB2 2QH, England.ms@mrc-lmb.cam.ac.uk