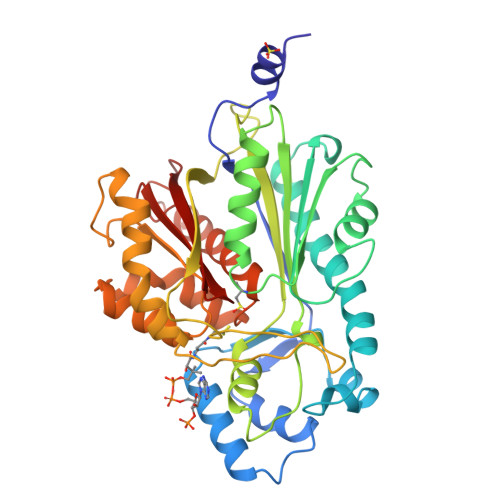
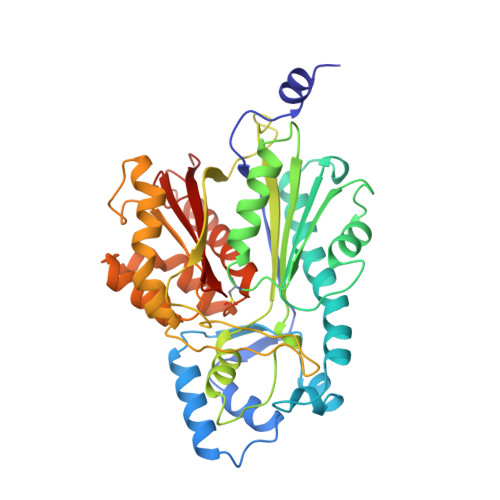
Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis.
Ferrer, J.L., Jez, J.M., Bowman, M.E., Dixon, R.A., Noel, J.P.(1999) Nat Struct Biol 6: 775-784
- PubMed: 10426957
- DOI: https://doi.org/10.1038/11553
- Primary Citation of Related Structures:
1BI5, 1BQ6, 1CGK, 1CGZ, 1CHW, 1CML - PubMed Abstract:
Chalcone synthase (CHS) is pivotal for the biosynthesis of flavonoid antimicrobial phytoalexins and anthocyanin pigments in plants. It produces chalcone by condensing one p-coumaroyl- and three malonyl-coenzyme A thioesters into a polyketide reaction intermediate that cyclizes. The crystal structures of CHS alone and complexed with substrate and product analogs reveal the active site architecture that defines the sequence and chemistry of multiple decarboxylation and condensation reactions and provides a molecular understanding of the cyclization reaction leading to chalcone synthesis. The structure of CHS complexed with resveratrol also suggests how stilbene synthase, a related enzyme, uses the same substrates and an alternate cyclization pathway to form resveratrol. By using the three-dimensional structure and the large database of CHS-like sequences, we can identify proteins likely to possess novel substrate and product specificity. The structure elucidates the chemical basis of plant polyketide biosynthesis and provides a framework for engineering CHS-like enzymes to produce new products.
Organizational Affiliation:
Structural Biology Laboratory, The Salk Institute for Biological Studies, 10010 N. Torrey Pines Rd., La Jolla, California 92037, USA.