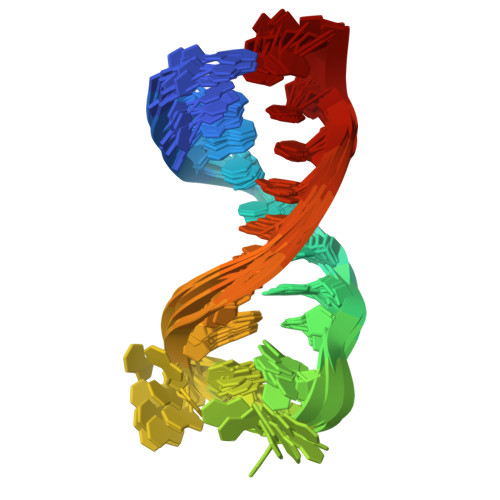
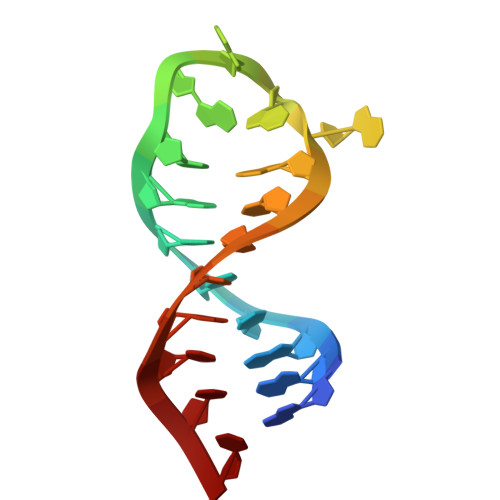
Three-dimensional folding of an RNA hairpin required for packaging HIV-1.
Pappalardo, L., Kerwood, D.J., Pelczer, I., Borer, P.N.(1998) J Mol Biol 282: 801-818
- PubMed: 9743628
- DOI: https://doi.org/10.1006/jmbi.1998.2046
- Primary Citation of Related Structures:
1BN0 - PubMed Abstract:
An NMR-based structure is presented for a 20 mer hairpin model of the SL3 stem-loop from the HIV-1 packaging signal. The stem has an A-family structure. However, the GGAG tetraloop appears to be flexible with the second (G10) and fourth (G12) bases extruded from the normal stacking arrangement. The A-base (A11) occupies a cavity large enough for it to jump rapidly between stacking upon G9 (in the loop) and G13 (from the base-pair adjacent to the loop). The H-bonding loci of G10, A11, and G12 are unoccupied in the free RNA structure. The loop should be easily adaptable to binding by the HIV-1 nucleocapsid protein or loop receptors.
Organizational Affiliation:
Department of Chemistry, Syracuse University, Syracuse, NY, 13244-4100, USA.