A novel complex of a phenolic derivative with insulin: structural features related to the T-->R transition.
Smith, G.D., Ciszak, E., Pangborn, W.(1996) Protein Sci 5: 1502-1511
- PubMed: 8844841
- DOI: https://doi.org/10.1002/pro.5560050806
- Primary Citation of Related Structures:
1BEN - PubMed Abstract:
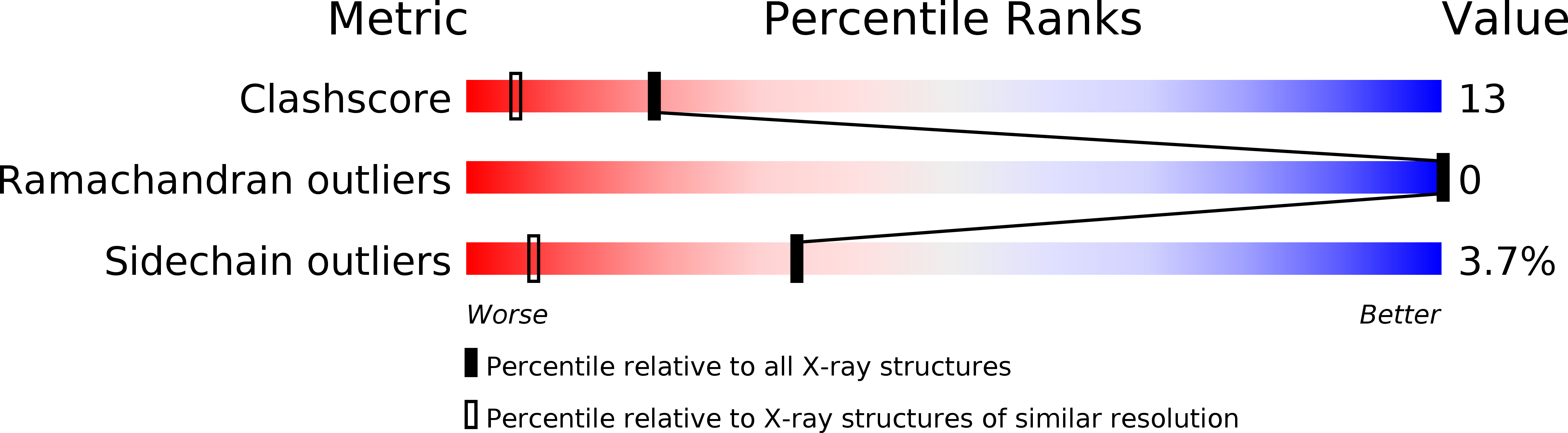
The structure of a symmetric T3R3f insulin hexamer, complexed with 4-hydroxybenzamide, has been determined using X-ray crystallographic techniques. Data were measured from six crystals grown in microgravity to a resolution of 1.4 A and the structure has been refined including the contributions from hydrogen atoms. The crystals are isomorphous with T3R3f complexes of phenolic derivatives as well as with uncomplexed forms. Unlike the structures of complexes with phenol, m-cresol, resorcinol, 4'-hydroxyacetanilide, and methylparaben, which bind one phenolic derivative molecule per R- or Rf-state monomer, two molecules of 4-hydroxybenzamide are bound by each Rf-state monomer. The presence of the second guest molecule results in an extensive hydrogen bonding network, mediated by water molecules, between the T- and Rf-state trimers and adds stability to the formation of the hexamer. The only access to these second sites is through three symmetry-related, narrow channels that originate on the surface of the T-state trimer. Although the conformation of the backbone atoms of the monomers is nearly identical to that of other T3R3f hexamers, significant changes are observed in the conformations of side chains in the vicinity of the second binding site. The side chain of the T-state A11 Cys residue, which forms a disulfide bond to A6 Cys in the same monomer, is observed in two discrete conformations; two discrete conformations are also present for the entire A8 Thr residue in the Rf-state monomer. A procedure is also described for an alternate method of interframe scaling and merging intensity data from an image plate detector.
Organizational Affiliation:
Hauptman-Woodward Medical Research Institute, Inc., Buffalo, New York 14203, USA. smith@hwi.buffalo.edu