Circular permutation of betaB2-crystallin changes the hierarchy of domain assembly.
Wright, G., Basak, A.K., Wieligmann, K., Mayr, E.M., Slingsby, C.(1998) Protein Sci 7: 1280-1285
- PubMed: 9655330
- DOI: https://doi.org/10.1002/pro.5560070602
- Primary Citation of Related Structures:
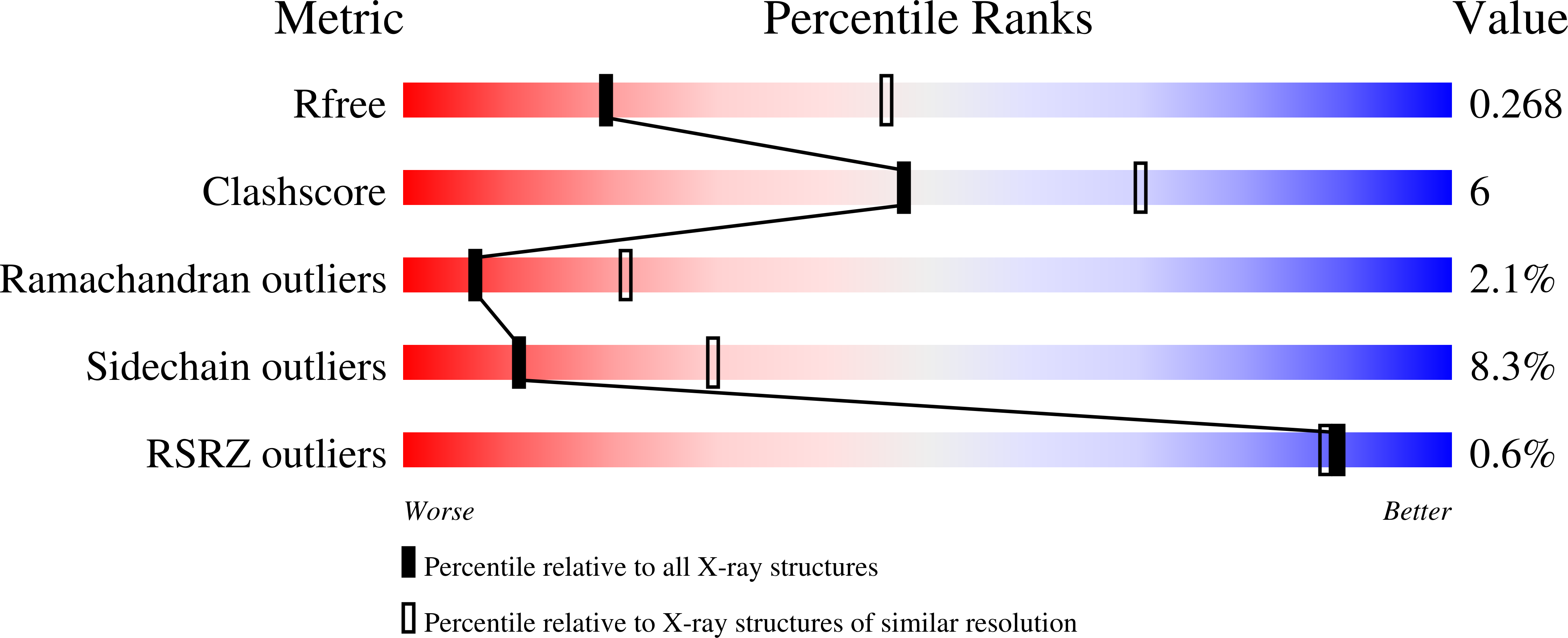
1BD7 - PubMed Abstract:
The betagamma-crystallins form a superfamily of eye lens proteins comprised of multiple Greek motifs that are symmetrically organized into domains and higher assemblies. In the betaB2-crystallin dimer each polypeptide folds into two similar domains that are related to monomeric gamma-crystallin by domain swapping. The crystal structure of the circularly permuted two-domain betaB2 polypeptide shows that permutation converts intermolecular domain pairing into intramolecular pairing. However, the dimeric permuted protein is, in fact, half a native tetramer. This result shows how the sequential order of domains in multi-domain proteins can affect quaternary domain assembly.
Organizational Affiliation:
Birkbeck College, Department of Crystallography, London, United Kingdom.