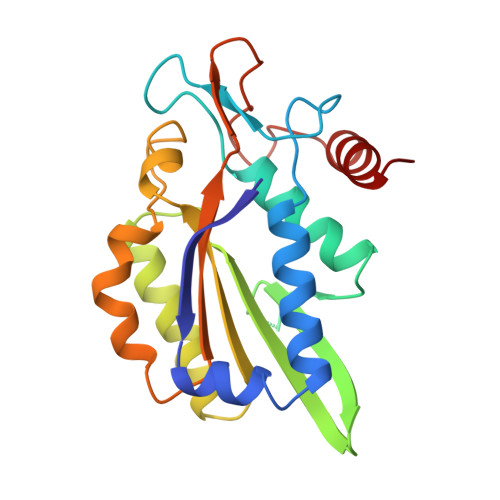
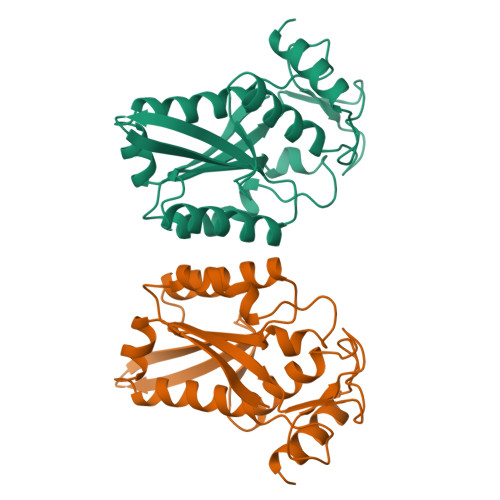
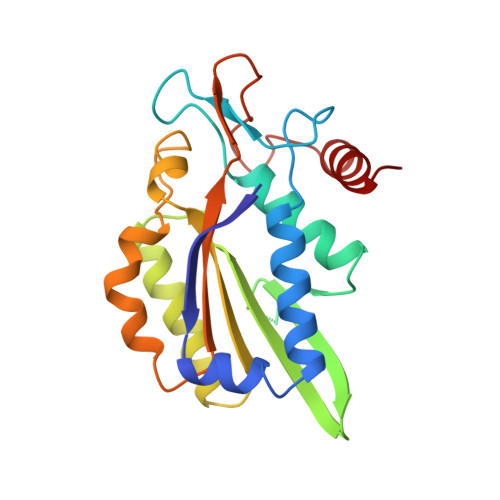
Structure of restriction endonuclease bamhi phased at 1.95 A resolution by MAD analysis.
Newman, M., Strzelecka, T., Dorner, L.F., Schildkraut, I., Aggarwal, A.K.(1994) Structure 2: 439-452
- PubMed: 8081758
- DOI: https://doi.org/10.1016/s0969-2126(00)00045-9
- Primary Citation of Related Structures:
1BAM - PubMed Abstract:
Type II restriction endonucleases recognize DNA sequences that vary between four to eight base pairs, and require only Mg2+ as a cofactor to catalyze the hydrolysis of DNA. Their protein sequences display a surprising lack of similarity, and no recurring structural motif analogous to the helix-turn-helix or the zinc finger of transcription factors, has yet been discovered. We have determined the crystal structure of restriction endonuclease BamHI at 1.95 A resolution. The structure was solved by combining phase information derived from multi-wavelength X-ray data by algebraic and maximum likelihood methods. The BamHI subunit consists of a central beta-sheet with alpha-helices on both sides. The dimer configuration reveals a large cleft which could accommodate B-form DNA. Mutants of the enzyme that are deficient in cleavage are located at or near the putative DNA-binding cleft. BamHI and endonuclease EcoRI share a common core motif (CCM) consisting of five beta-strands and two helices. It remains to be determined if other restriction enzymes also contain the CCM. The structure of BamHI provides the first clear evidence that there may be substantial structural homology amongst restriction enzymes, even though it is undetectable at the sequence level.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY 10032.