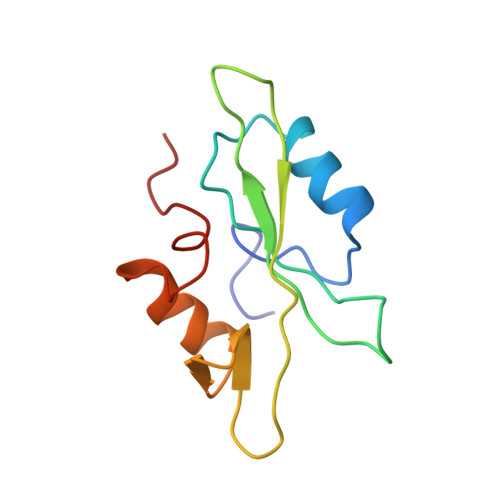
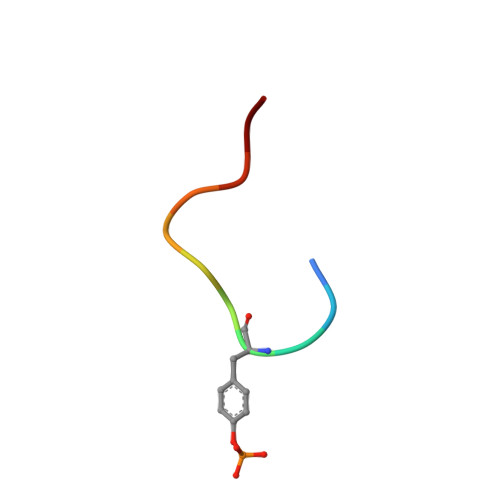
The SH2 domain from the tyrosine kinase Fyn in complex with a phosphotyrosyl peptide reveals insights into domain stability and binding specificity.
Mulhern, T.D., Shaw, G.L., Morton, C.J., Day, A.J., Campbell, I.D.(1997) Structure 5: 1313-1323
- PubMed: 9351806
- DOI: https://doi.org/10.1016/s0969-2126(97)00283-9
- Primary Citation of Related Structures:
1AOT, 1AOU - PubMed Abstract:
SH2 domains are found in a variety of signal transduction proteins; they bind phosphotyrosine-containing sequences, allowing them to both recognize target molecules and regulate intramolecular kinase activity. Fyn is a member of the Src family of tyrosine kinases that are involved in signal transduction by association with a number of membrane receptors. The kinase activity of these signalling proteins is modulated by switching the binding mode of their SH2 and SH3 domains from intramolecular to intermolecular. The molecular basis of the signalling roles observed for different Src family members is still not well understood; although structures have been determined for the SH2 domains of other Src family molecules, this is the first structure of the Fyn SH2 domain. The structure of the Fyn SH2 domain in complex with a phosphotyrosyl peptide (EPQpYEEIPIYL) was determined by high resolution NMR spectroscopy. The overall structure of the complex is analogous to that of other SH2-peptide complexes. Noteworthy aspects of the structure are: the BG loop, which contacts the bound peptide, contains a type-I' turn; a capping-box-like interaction is present at the N-terminal end of helix alpha A; cis-trans isomerization of the Val beta G1-Pro beta G2 peptide bond causes conformational heterogeneity of residues near the N and C termini of the domain. Comparison of the Fyn SH2 domain structure with other structures of SH2 domains highlights several interesting features. Conservation of helix capping interactions among various SH2 domains is suggestive of a role in protein stabilisation. The presence of a type-I' turn in the BG loop, which is dependent on the presence of a glycine residue at position BG3, is indicative of a binding pocket, characteristic of the Src family, SykC and Abl, rather than a binding groove found in PLC-gamma 1C, p85 alpha N and Shc, for example.
Organizational Affiliation:
Oxford Centre for Molecular Sciences, UK.