Biochemical and crystallographic analyses of a portal mutant of the adipocyte lipid-binding protein.
Ory, J., Kane, C.D., Simpson, M.A., Banaszak, L.J., Bernlohr, D.A.(1997) J Biological Chem 272: 9793-9801
- PubMed: 9092513
- DOI: https://doi.org/10.1074/jbc.272.15.9793
- Primary Citation of Related Structures:
1AB0, 1ACD - PubMed Abstract:
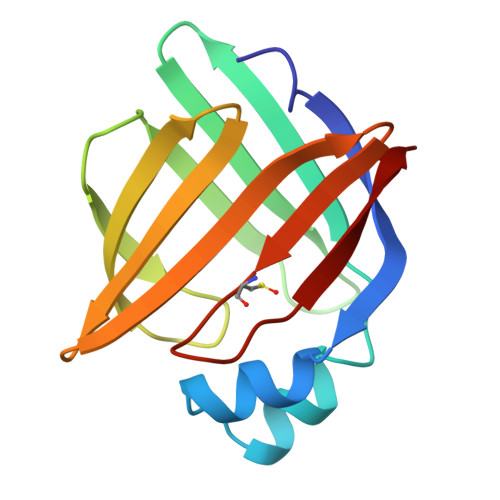
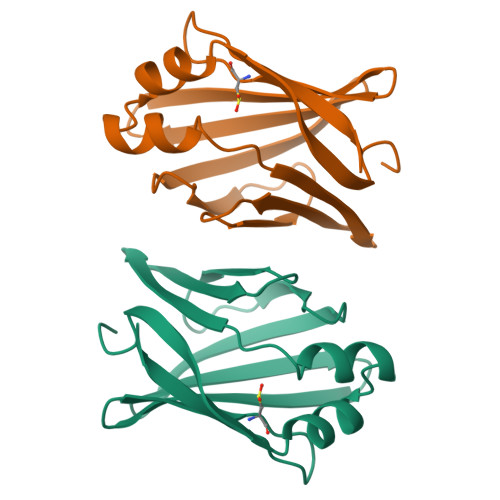
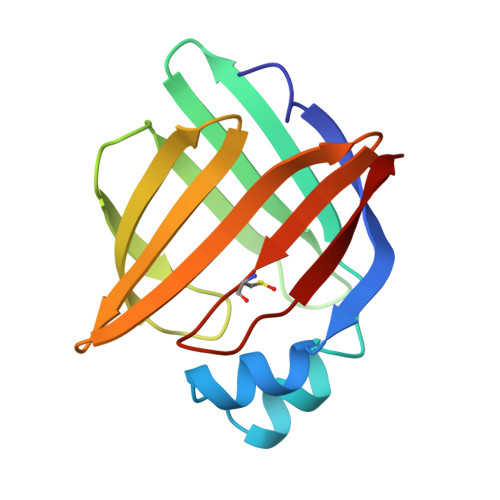
A number of crystallographic studies of the adipocyte lipid-binding protein have established that the fatty acid-binding site is within an internalized water-filled cavity. The same studies have also suggested the existence of a region physically distinct from the fatty acid-binding site which connects the cavity of the protein with the external solvent, hereafter referred to as the portal. In an effort to examine the portal region, we have used site-directed mutagenesis to introduce the mutations V32D/F57H into the murine ALBP cDNA. Mutant protein has been isolated, crystallized, and its stability and binding properties studied by biochemical methods. As assessed by guanidine-HCl denaturation, the mutant form exhibited a slight overall destabilization relative to the wild-type protein under both acid and alkaline conditions. Accessibility to the cavity in both the mutant and wild-type proteins was observed by stopped-flow analysis of the modification of a cavity residue, Cys117, by the sulfhydryl reactive agent 5, 5'-dithiobis(2-nitrobenzoic acid) at pH 8.5. Cys117 of V32D/F57H ALBP was modified 7-fold faster than the wild-type protein. The ligand binding properties of both the V32D/F57H mutant and wild-type proteins were analyzed using a fluorescent probe at pH 6.0 and 8.0. The apparent dissociation constants for 1-anilinonaphthalene-8-sulfonic acid were approximately 9-10-fold greater than the wild-type protein, independent of pH. In addition, there is a 6-fold increase in the Kd for oleic acid for the portal mutant relative to the wild-type at pH 8.0. To study the effect of pH on the double mutant, it was crystallized and analyzed in two distinct space groups at pH 4.5 and 6.4. While in general the differences in the overall main chain conformations are negligible, changes were observed in the crystallographic structures near the site of the mutations. At both pH values, the mutant side chains are positioned somewhat differently than in wild-type protein. To ensure that the mutations had not altered ionic conditions near the binding site, the crystallographic coordinates were used to monitor the electrostatic potentials from the head group site to the positions near the portal region. The differences in the electrostatic potentials were small in all regions, and did not explain the differences in ligand affinity. We present these results within the context of fatty acid binding and suggest lipid association is more complex than that described within a single equilibrium event.
- Department of Biochemistry, University of Minnesota, St. Paul, Minnesota 55108, USA.
Organizational Affiliation: