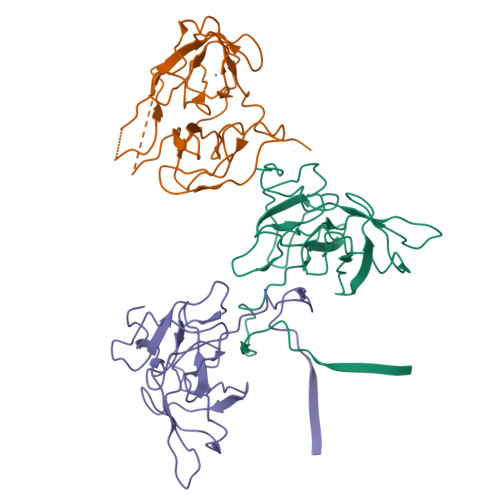
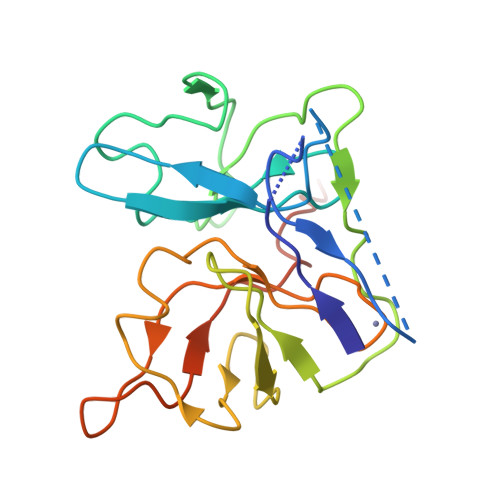
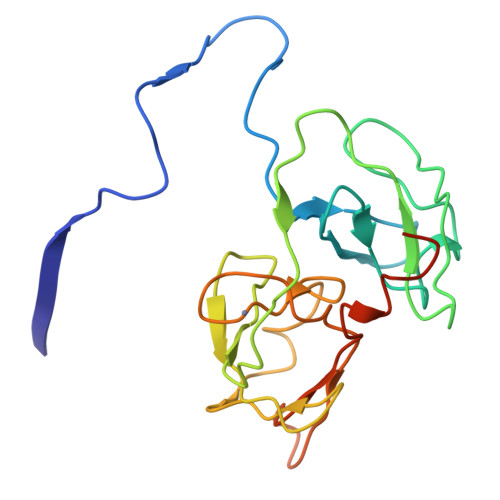
The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site.
Love, R.A., Parge, H.E., Wickersham, J.A., Hostomsky, Z., Habuka, N., Moomaw, E.W., Adachi, T., Hostomska, Z.(1996) Cell 87: 331-342
- PubMed: 8861916
- DOI: https://doi.org/10.1016/s0092-8674(00)81350-1
- Primary Citation of Related Structures:
1A1Q - PubMed Abstract:
During replication of hepatitis C virus (HCV), the final steps of polyprotein processing are performed by a viral proteinase located in the N-terminal one-third of nonstructural protein 3. The structure of NS3 proteinase from HCV BK strain was determined by X-ray crystallography at 2.4 angstrom resolution. NS3P folds as a trypsin-like proteinase with two beta barrels and a catalytic triad of His-57, Asp-81, Ser-139. The structure has a substrate-binding site consistent with the cleavage specificity of the enzyme. Novel features include a structural zinc-binding site and a long N-terminus that interacts with neighboring molecules by binding to a hydrophobic surface patch.
Organizational Affiliation:
Agouron Pharmaceuticals, Inc., San Diego, California 92121, USA.