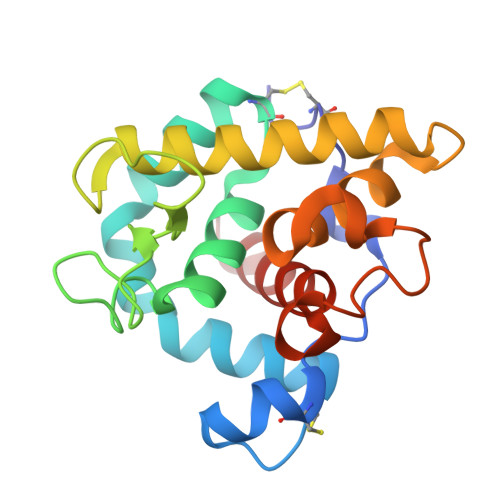
The refined structures of goose lysozyme and its complex with a bound trisaccharide show that the "goose-type" lysozymes lack a catalytic aspartate residue.
Weaver, L.H., Grutter, M.G., Matthews, B.W.(1995) J Mol Biol 245: 54-68
- PubMed: 7823320
- DOI: https://doi.org/10.1016/s0022-2836(95)80038-7
- Primary Citation of Related Structures:
153L, 154L - PubMed Abstract:
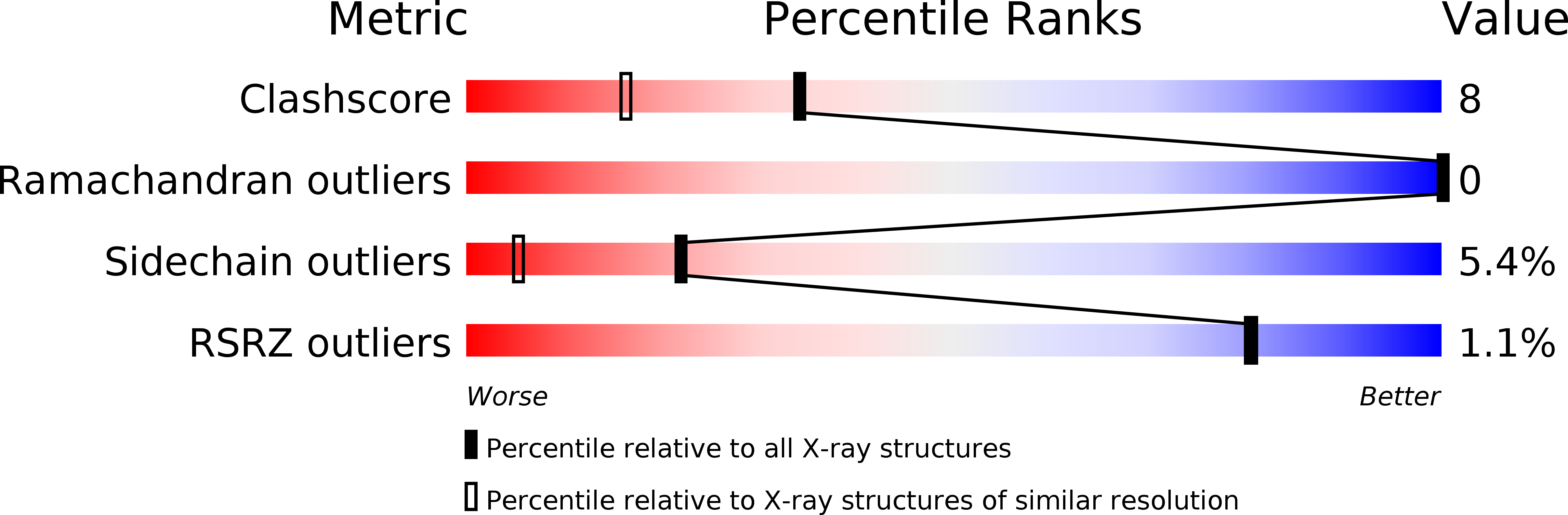
The structure of goose egg-white lysozyme (GEWL) has been refined to an R-value of 15.9% at 1.6 A resolution. Details of the structure determination, the refinement and the structure itself are presented. The structure of a complex of the enzyme with the trisaccharide of N-acetyl glucosamine has also been determined and refined at 1.6 A resolution. The trisaccharide occupies sites analogous to the B, C and D subsites of chicken (HEWL) and phage T4 (T4L) lysozymes. All three lysozymes (GEWL, HEWL and T4L) display the same characteristic set of bridging hydrogen bonds between backbone atoms of the protein and the 2-acetamido group of the saccharide in subsite C. Glu73 of GEWL is seen to correspond closely to Glu35 of HEWL (and to Glu11 of T4L) and supports the established view that this group is critically involved in the catalytic mechanism. There is, however, no obvious residue in goose lysozyme that is a counterpart of Asp52 of chicken lysozyme (or of Asp20 in T4L), suggesting that a second acidic residue is not essential for the catalytic activity of goose lysozyme, and may not be required for the activity of other lysozymes.
Organizational Affiliation:
Institute of Molecular Biology, Howard Hughes Medical Institute, Eugene, OR.