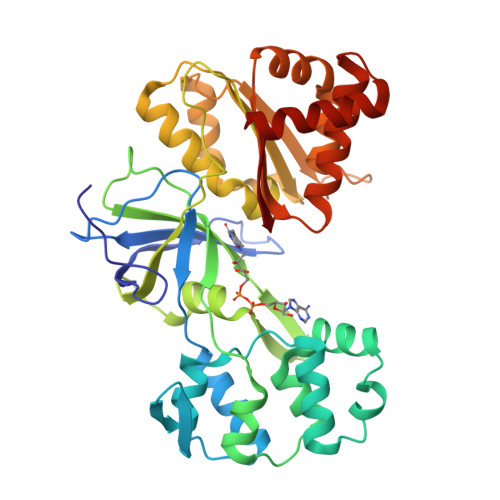
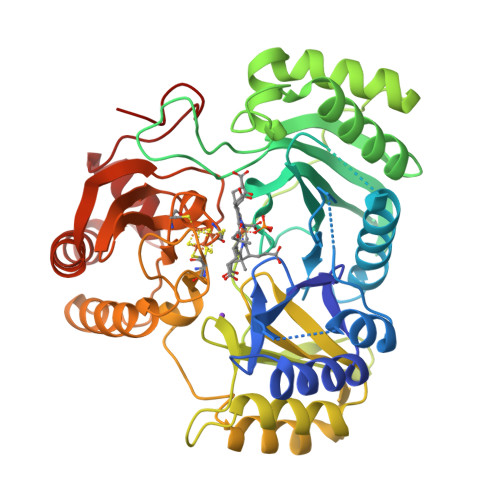
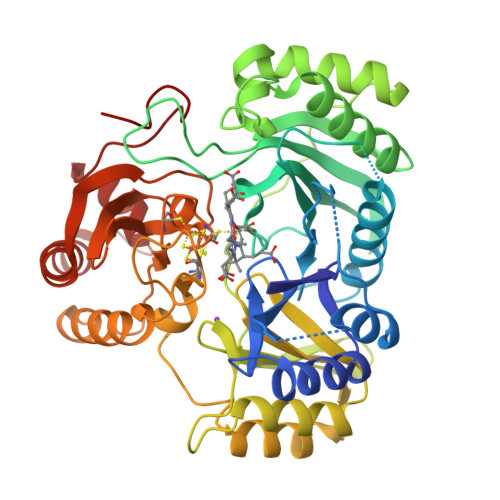
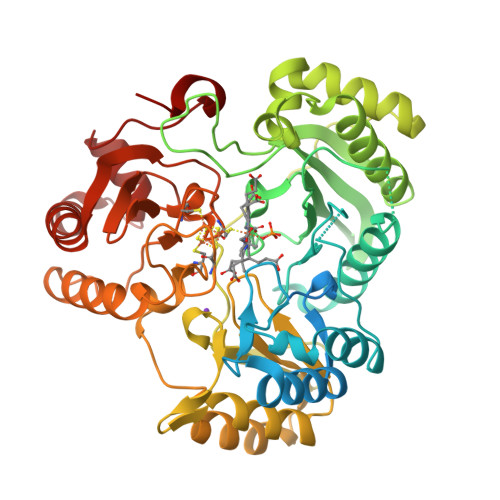
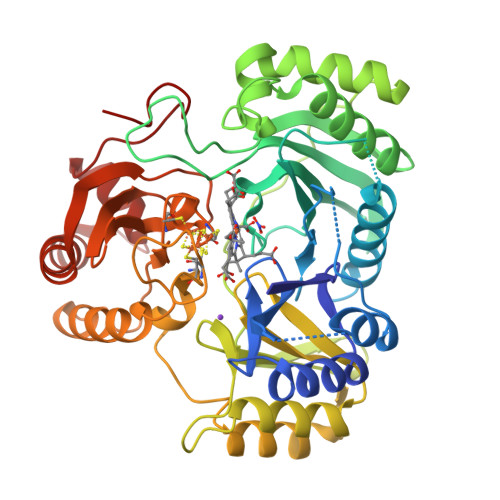
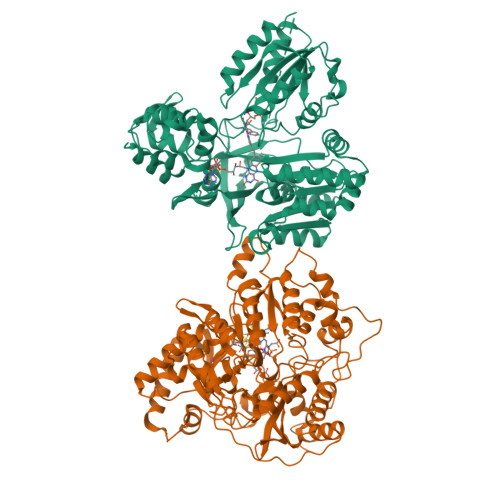
Structure Determination MethodologyScientific Name of Source OrganismRefinement Resolution (Å)Enzyme Classification Name | Gruez, A., Pignol, D., Zeghouf, M., Coves, J., Fontecave, M., Ferrer, J.L., Fontecilla-Camps, J.C. (2000) J Mol Biology 299: 199-212 | Released | 2000-11-13 | | Method | X-RAY DIFFRACTION 2.01 Å | | Organisms | | | Macromolecule | | | Unique Ligands | FAD, SO4 |
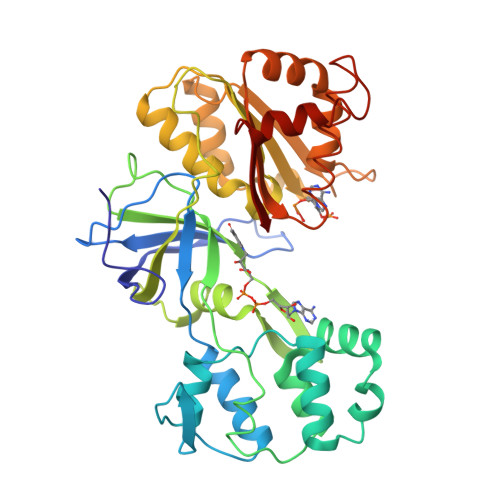
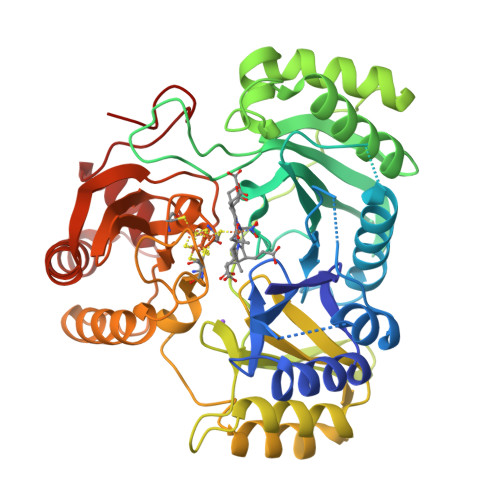
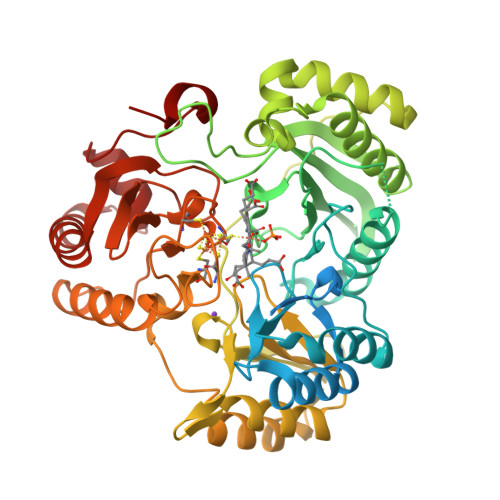
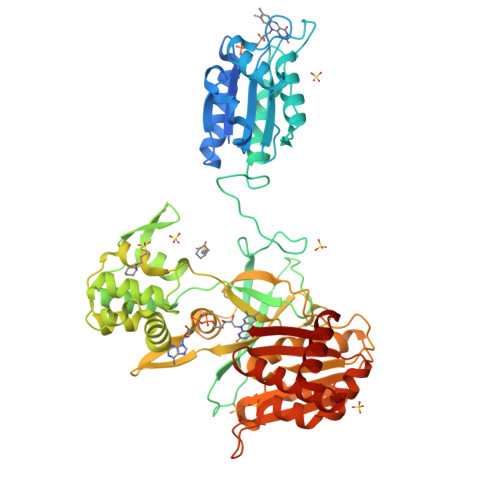
Gruez, A., Pignol, D., Zeghouf, M., Coves, J., Fontecave, M., Ferrer, J.L., Fontecilla-Camps, J.C. (2000) J Mol Biology 299: 199-212 | Released | 2000-11-13 | | Method | X-RAY DIFFRACTION 2.51 Å | | Organisms | | | Macromolecule | | | Unique Ligands | FAD, NAP |
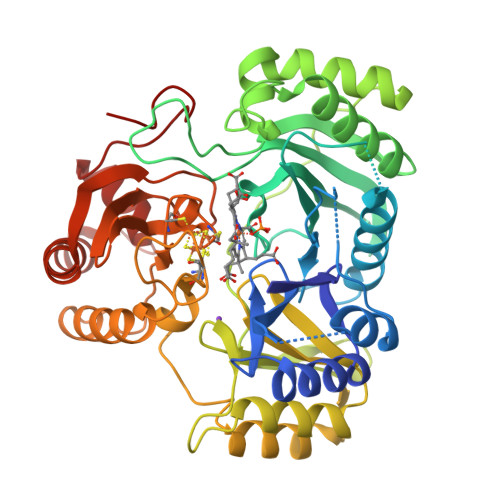
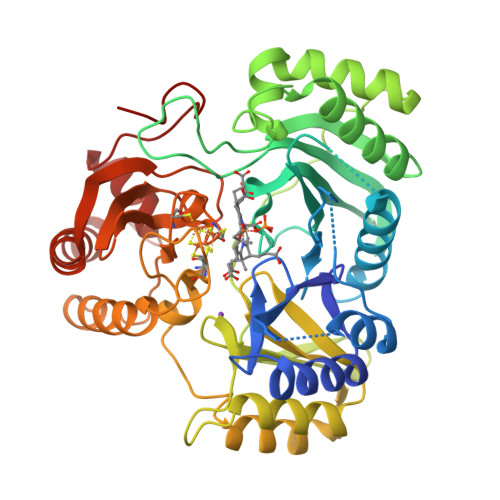
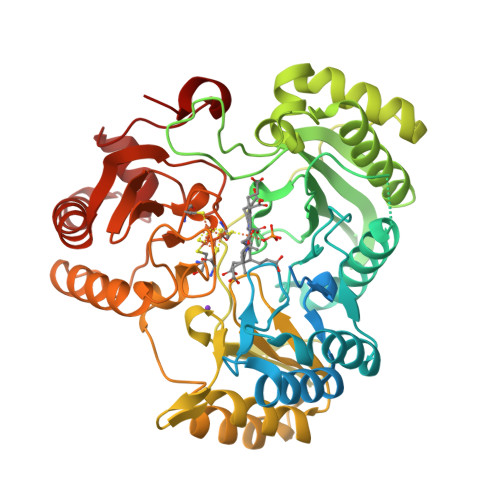
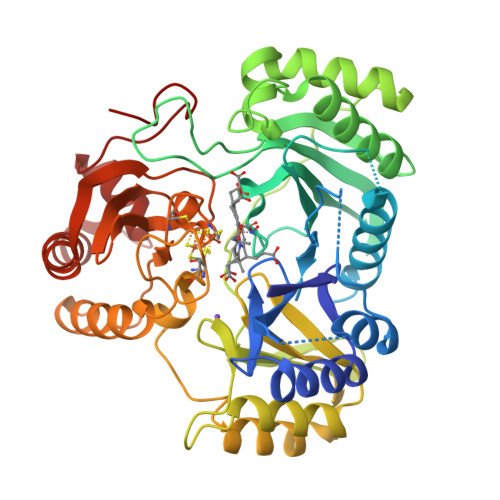
Crane, B.R., Getzoff, E.D. (1997) Biochemistry 36: 12101-12119 | Released | 1998-01-14 | | Method | X-RAY DIFFRACTION 2.2 Å | | Organisms | | | Macromolecule | | | Unique Ligands | K, SF4, SRM |
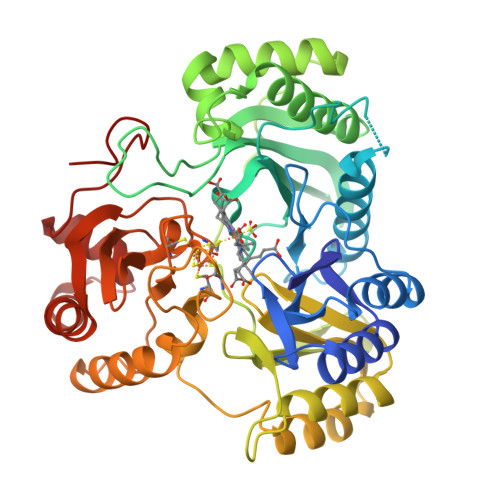
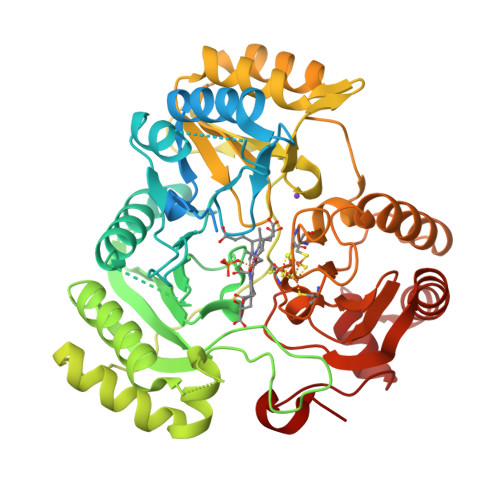
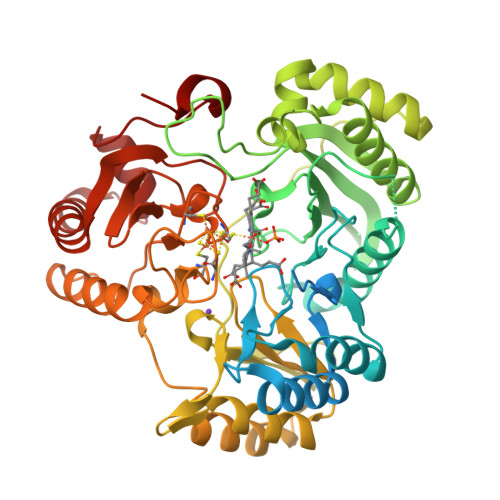
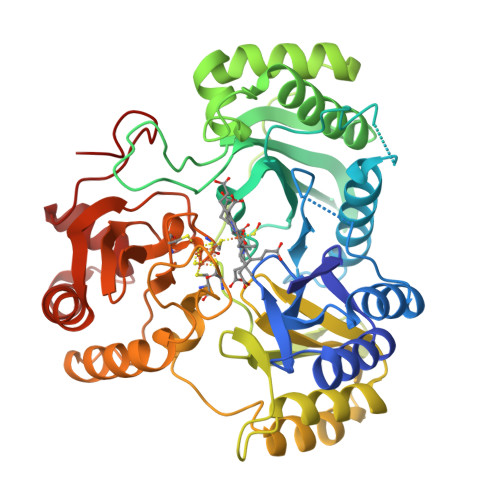
Stroupe, M.E. (2018) Biochim Biophys Acta 1866: 933-940 | Released | 2018-06-13 | | Method | X-RAY DIFFRACTION 1.5 Å | | Organisms | | | Macromolecule | | | Unique Ligands | K, PO4, SF4, SRM |
Stroupe, M.E. (2018) Biochim Biophys Acta 1866: 933-940 | Released | 2018-06-13 | | Method | X-RAY DIFFRACTION 1.542 Å | | Organisms | | | Macromolecule | | | Unique Ligands | K, PO4, SF4, SRM |
Stroupe, M.E. (2018) Biochim Biophys Acta 1866: 933-940 | Released | 2018-06-13 | | Method | X-RAY DIFFRACTION 1.542 Å | | Organisms | | | Macromolecule | | | Unique Ligands | K, PO4, SF4, SRM |
Stroupe, M.E. (2018) Biochim Biophys Acta 1866: 933-940 | Released | 2018-06-13 | | Method | X-RAY DIFFRACTION 1.681 Å | | Organisms | | | Macromolecule | | | Unique Ligands | K, PO4, SF4, SRM |
Tavolieri, A.M., Askenasy, I., Murray, D.T., Pennington, J.M., Stroupe, M.E. (2019) J Struct Biol 205: 170-179 | Released | 2019-02-27 | | Method | X-RAY DIFFRACTION 2.341 Å | | Organisms | | | Macromolecule | | | Unique Ligands | CXS, FAD, FMN, SO4 |
Ghazi Esfahani, B., Walia, N., Neselu, K., Aragon, M., Askenasy, I., Wei, A., Mendez, J.H., Stroupe, M.E. To be published | Released | 2025-02-12 | | Method | ELECTRON MICROSCOPY 2.78 Å | | Organisms | | | Macromolecule | | | Unique Ligands | FAD, FMN, K, PO4, SF4, SRM |
|