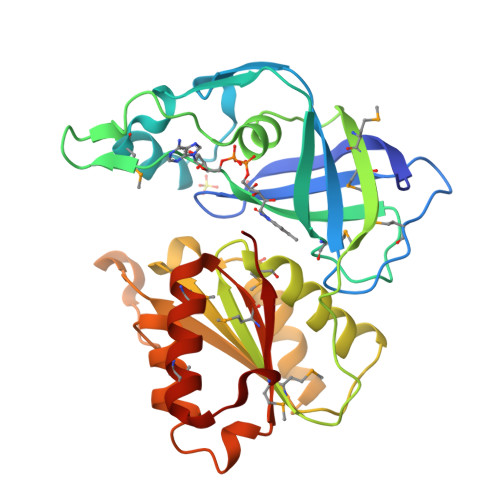
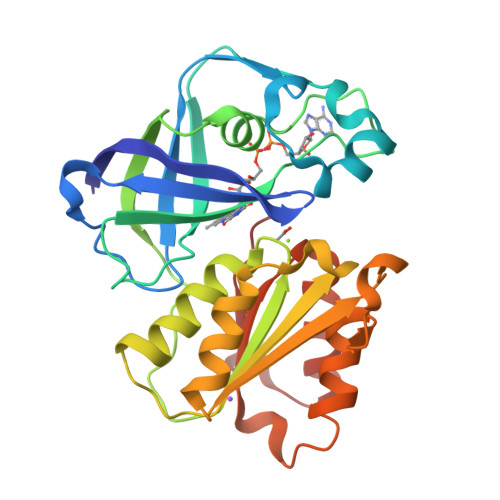
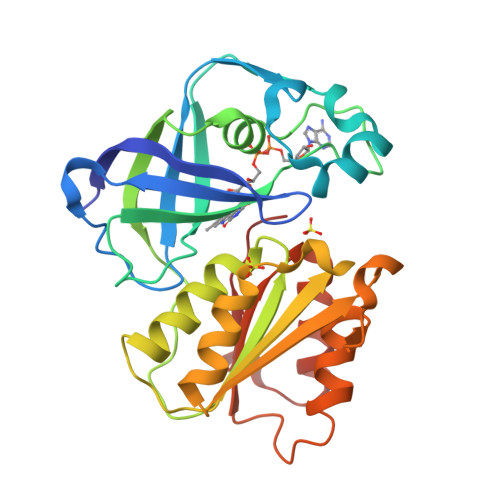
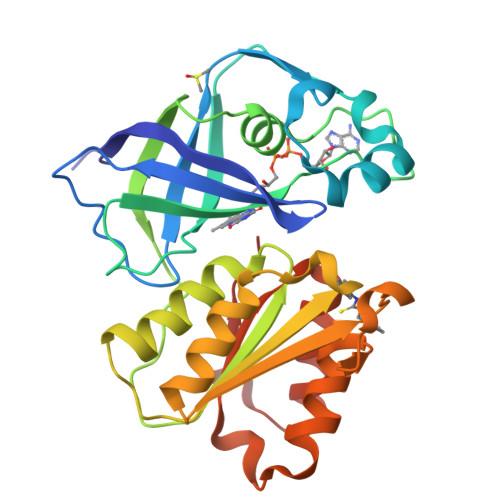
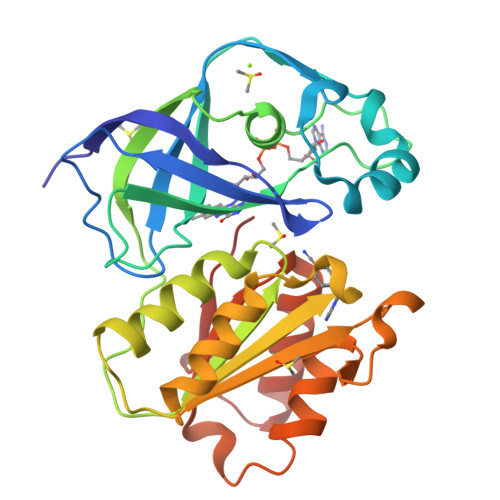
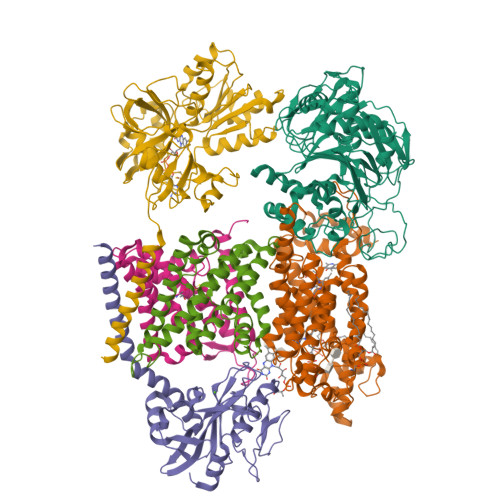
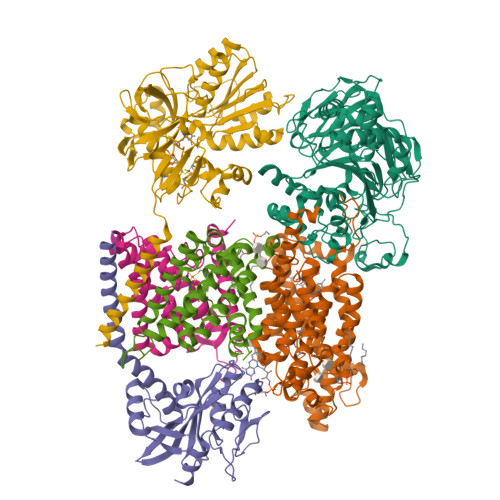
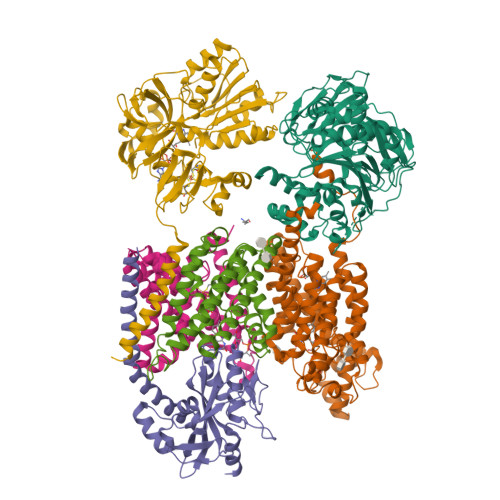
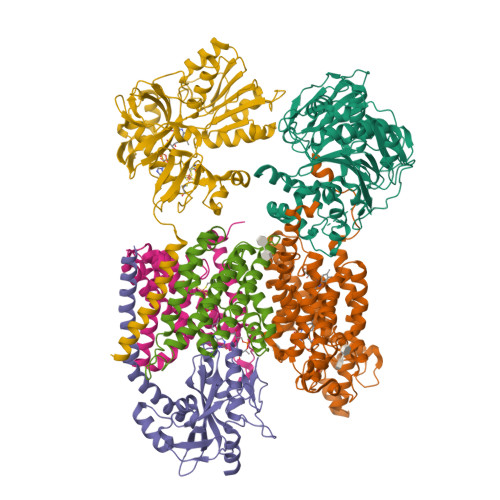
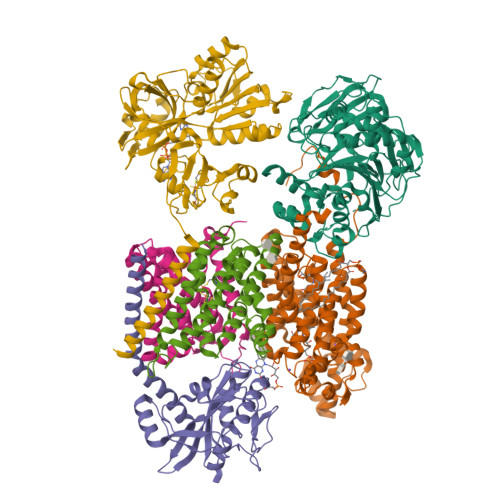
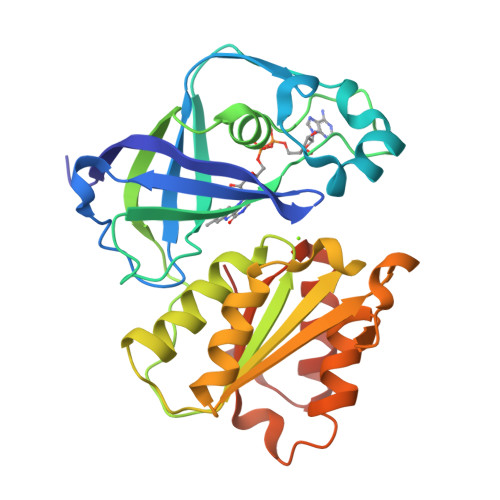
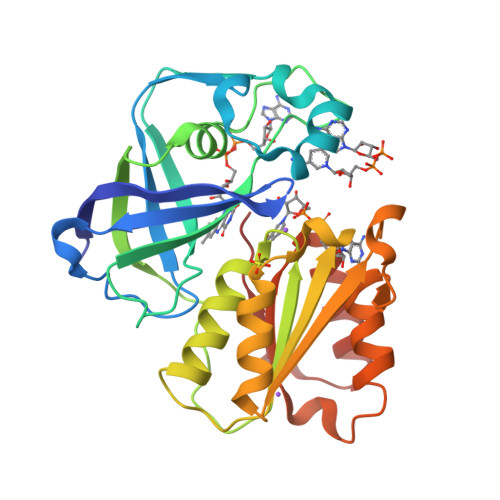
Structure Determination MethodologyScientific Name of Source OrganismRefinement Resolution (Å)Enzyme Classification NameMembrane Protein Annotation | Fritz, G. (2014) Nature 516: 62-67 | Released | 2014-12-03 | | Method | X-RAY DIFFRACTION 1.55 Å | | Organisms | | | Macromolecule | | | Unique Ligands | ACT, FAD, MG, NA |
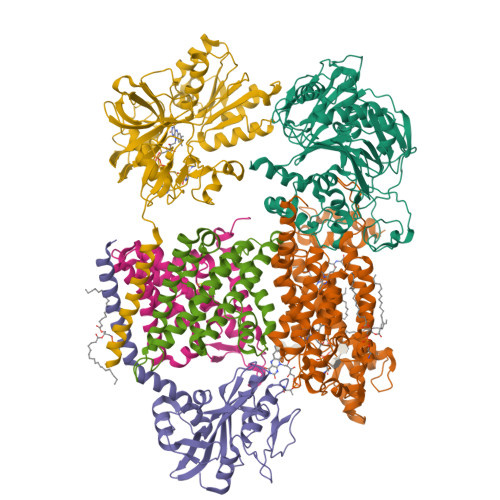
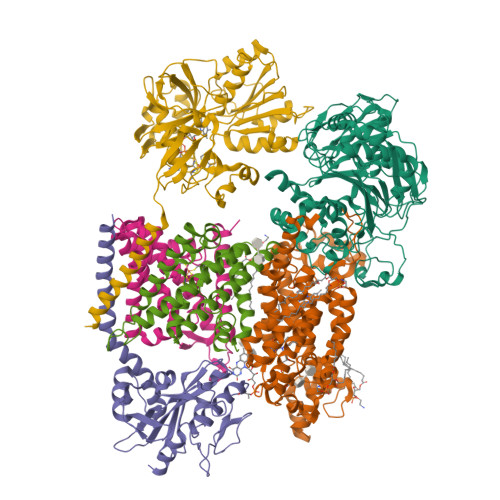
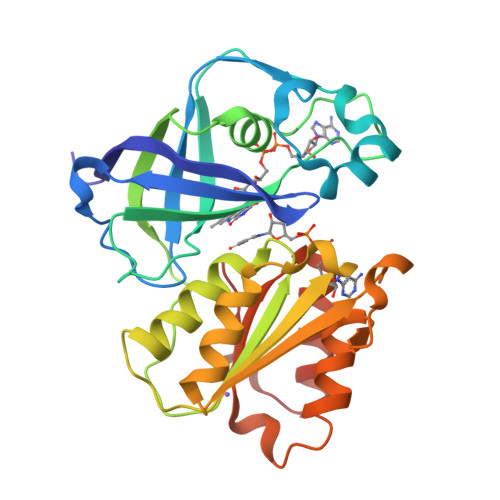
Fritz, G. (2014) Nature 516: 62-67 | Released | 2015-08-05 | | Method | X-RAY DIFFRACTION 2.7019 Å | | Organisms | | | Macromolecule | | | Unique Ligands | FAD, SO4 |
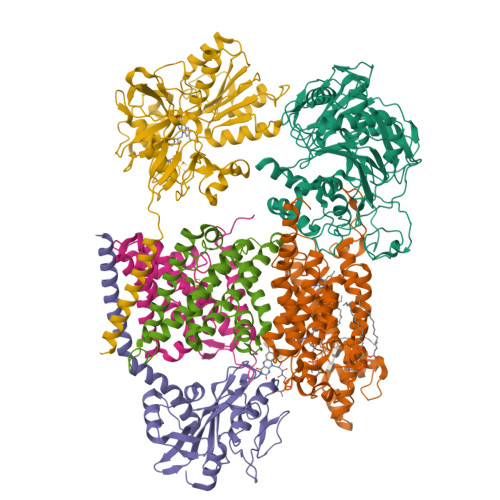
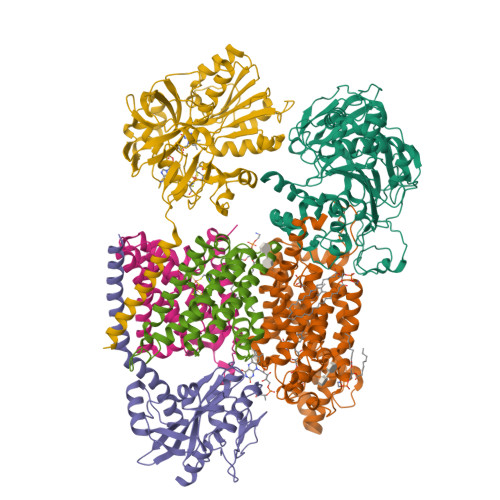
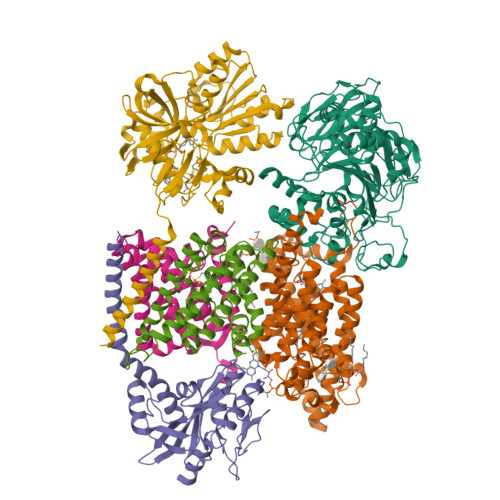
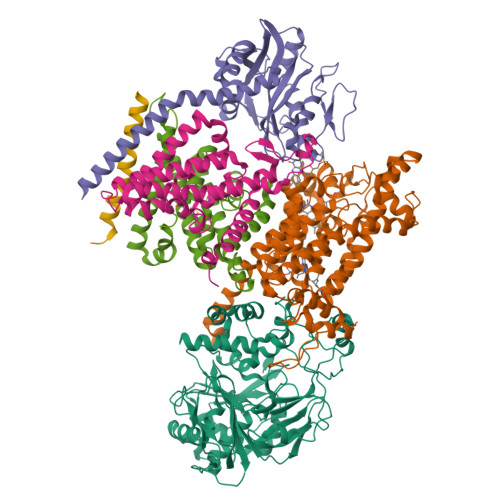
Kishikawa, J., Ishikawa, M., Masuya, T., Murai, M., Barquera, B., Miyoshi, H. (2022) Nat Commun 13: 4082-4082 | Released | 2022-07-20 | | Method | ELECTRON MICROSCOPY 3.1 Å | | Organisms | | | Macromolecule | Unique protein chains: 6 | | Unique Ligands | CA, FAD, FES, FMN, LMT, PEE, RBF |
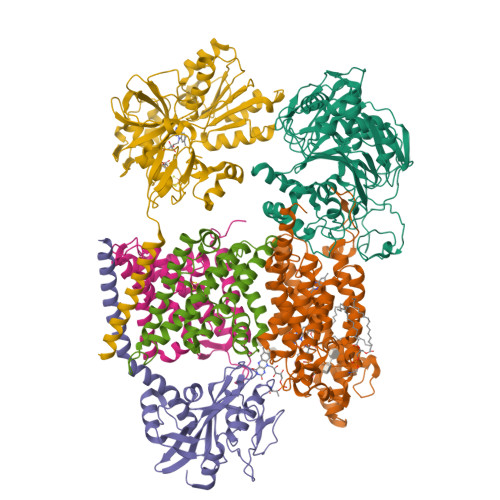
Kishikawa, J., Ishikawa, M., Masuya, T., Murai, M., Barquera, B., Miyoshi, H. (2022) Nat Commun 13: 4082-4082 | Released | 2022-07-20 | | Method | ELECTRON MICROSCOPY 3.1 Å | | Organisms | | | Macromolecule | Unique protein chains: 6 | | Unique Ligands | CA, FAD, FES, FMN, LMT, PEE, RBF |
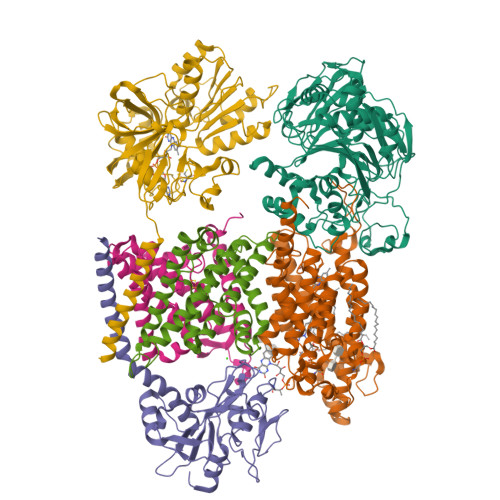
Kishikawa, J., Ishikawa, M., Masuya, T., Murai, M., Barquera, B., Miyoshi, H. (2022) Nat Commun 13: 4082-4082 | Released | 2022-07-20 | | Method | ELECTRON MICROSCOPY 3.1 Å | | Organisms | | | Macromolecule | Unique protein chains: 6 | | Unique Ligands | CA, FAD, FES, FMN, LMT, PEE, RBF |
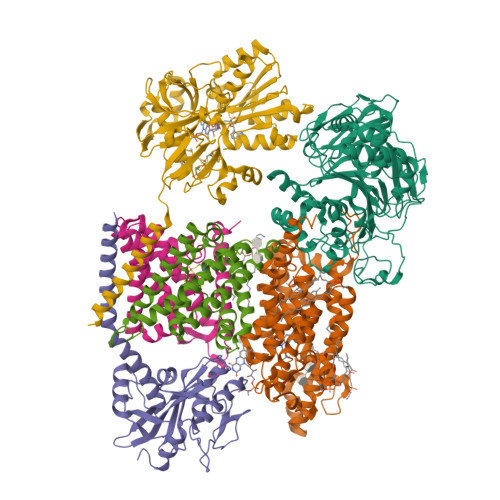
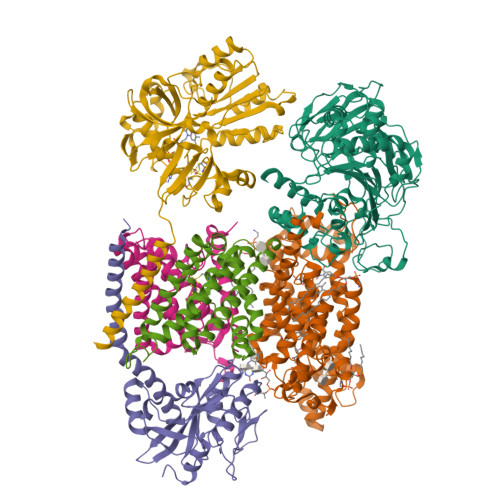
Kishikawa, J., Ishikawa, M., Masuya, T., Murai, M., Barquera, B., Miyoshi, H. (2022) Nat Commun 13: 4082-4082 | Released | 2022-07-20 | | Method | ELECTRON MICROSCOPY 3 Å | | Organisms | | | Macromolecule | Unique protein chains: 6 | | Unique Ligands | 0NI, CA, FAD, FES, FMN, LMT, PEE, RBF |
Kishikawa, J., Ishikawa, M., Masuya, T., Murai, M., Barquera, B., Miyoshi, H. (2022) Nat Commun 13: 4082-4082 | Released | 2022-07-20 | | Method | ELECTRON MICROSCOPY 2.9 Å | | Organisms | | | Macromolecule | Unique protein chains: 6 | | Unique Ligands | CA, FAD, FES, FMN, IQT, LMT, PEE, RBF |
Hau, J.-L., Kaltwasser, S., Vonck, J., Fritz, G., Steuber, J. (2023) Nat Struct Mol Biol 30: 1686-1694 | Released | 2023-06-14 | | Method | ELECTRON MICROSCOPY 3.37 Å | | Organisms | | | Macromolecule | Unique protein chains: 6 | | Unique Ligands | 3PE, FAD, FES, FMN, K, LMT, NA, RBF |
Hau, J.-L., Kaltwasser, S., Vonck, J., Fritz, G., Steuber, J. (2023) Nat Struct Mol Biol 30: 1686-1694 | Released | 2023-09-20 | | Method | ELECTRON MICROSCOPY 2.86 Å | | Organisms | | | Macromolecule | Unique protein chains: 6 | | Unique Ligands | 3PE, FAD, FES, FMN, LMT, NA, NAI, RBF, UQ2 |
Hau, J.-L., Kaltwasser, S., Vonck, J., Fritz, G., Steuber, J. (2023) Nat Struct Mol Biol 30: 1686-1694 | Released | 2023-06-14 | | Method | ELECTRON MICROSCOPY 2.73 Å | | Organisms | | | Macromolecule | Unique protein chains: 6 | | Unique Ligands | 3PE, FAD, FES, FMN, LMT, NA, RBF, UQ2 |
Hau, J.-L., Kaltwasser, S., Vonck, J., Fritz, G., Steuber, J. (2023) Nat Struct Mol Biol 30: 1686-1694 | Released | 2023-06-14 | | Method | ELECTRON MICROSCOPY 2.56 Å | | Organisms | | | Macromolecule | Unique protein chains: 6 | | Unique Ligands | 3PE, FAD, FES, FMN, LMT, NA, RBF, UQ1 |
Hau, J.-L., Kaltwasser, S., Vonck, J., Fritz, G., Steuber, J. (2023) Nat Struct Mol Biol 30: 1686-1694 | Released | 2023-06-14 | | Method | ELECTRON MICROSCOPY 3.2 Å | | Organisms | | | Macromolecule | Unique protein chains: 6 | | Unique Ligands | 3PE, FAD, FES, FMN, LMT, RBF |
Hau, J.-L., Kaltwasser, S., Vonck, J., Fritz, G., Steuber, J. (2023) Nat Struct Mol Biol 30: 1686-1694 | Released | 2023-06-14 | | Method | ELECTRON MICROSCOPY 3.3 Å | | Organisms | | | Macromolecule | Unique protein chains: 6 | | Unique Ligands | 3PE, FAD, FES, FMN, HQO, LMT, RBF |
Fritz, G. (2023) Nat Struct Mol Biol 30: 1686-1694 | Released | 2023-07-12 | | Method | X-RAY DIFFRACTION 3.4 Å | | Organisms | | | Macromolecule | Unique protein chains: 6 | | Unique Ligands | FAD, FES, FMN, K, LMT, LYS, RBF |
Fritz, G. (2023) Nat Struct Mol Biol 30: 1686-1694 | Released | 2023-07-12 | | Method | X-RAY DIFFRACTION 3.5 Å | | Organisms | | | Macromolecule | Unique protein chains: 6 | | Unique Ligands | FAD, FES, FMN, LMT, NA, RBF |
Fritz, G. (2023) Nat Struct Mol Biol | Released | 2023-07-12 | | Method | X-RAY DIFFRACTION 3.11 Å | | Organisms | | | Macromolecule | Unique protein chains: 6 | | Unique Ligands | 3PE, BR, FAD, FES, FMN, K, LMT, NA, RBF |
Fritz, G. (2023) Nat Struct Mol Biol 30: 1686-1694 | Released | 2023-07-12 | | Method | X-RAY DIFFRACTION 1.55 Å | | Organisms | | | Macromolecule | | | Unique Ligands | FAD, MG |
Fritz, G. (2023) Nat Struct Mol Biol 30: 1686-1694 | Released | 2023-07-12 | | Method | X-RAY DIFFRACTION 1.5 Å | | Organisms | | | Macromolecule | | | Unique Ligands | FAD, NA, NAI, SO4 |
Fritz, G. (2023) Nat Struct Mol Biol 30: 1686-1694 | Released | 2023-07-12 | | Method | X-RAY DIFFRACTION 1.65 Å | | Organisms | | | Macromolecule | | | Unique Ligands | FAD, NA, NAI |
Fuller, J.R., Juarez, O. To be published | Released | 2022-11-16 | | Method | ELECTRON MICROSCOPY 2.5804 Å | | Organisms | | | Macromolecule | Unique protein chains: 6 | | Unique Ligands | FES, FMN, RBF, UQ1 |
|