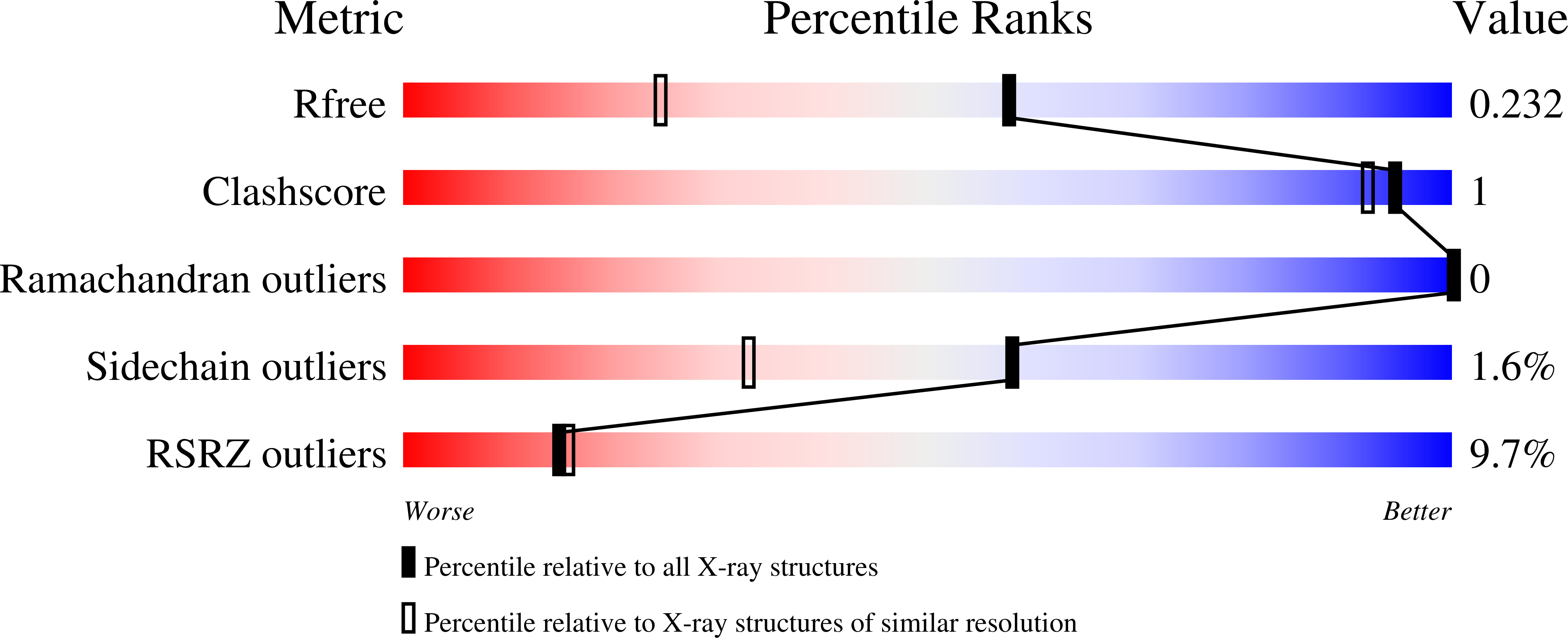
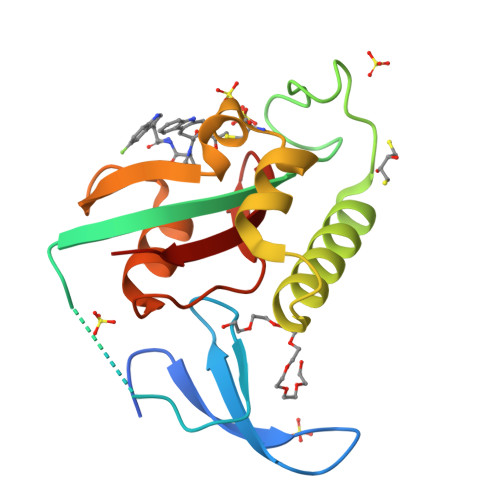
Targeted degradation of Pin1 by protein-destabilizing compounds.
Alboreggia, G., Udompholkul, P., Rodriguez, I., Blaha, G., Pellecchia, M.(2024) Proc Natl Acad Sci U S A 121: e2403330121-e2403330121
- PubMed: 39531501
- DOI: https://doi.org/10.1073/pnas.2403330121
- Primary Citation of Related Structures:
8VJD, 8VJE, 8VJF, 8VJG - PubMed Abstract:
The concept of targeted protein degradation is at the forefront of modern drug discovery, which aims to eliminate disease-causing proteins using specific molecules. In this paper, we explored the idea to design protein degraders based on the section of ligands that cause protein destabilization, hence that facilitate the cellular breakdown of the target. Our studies present covalent agents targeting Pin1, a cis-trans prolyl isomerase that plays a crucial role in tumorigenesis. Our design strategy entailed iterative optimizations of agents for potency and Pin1 destabilization in vitro. Biophysical and cellular studies suggest that the agents may act like molecular crowbars , displacing protein-stabilizing interactions that open the structure for recognition by the proteasome degradation machinery. This approach resulted in a series of potent and effective Pin1 degraders with potential applications in target validation and in therapeutic development. We propose that our design strategy can identify molecular degraders without engineering bifunctional agents that artificially create interactions between a disease-causing protein and a ubiquitin ligase.
Organizational Affiliation:
Division of Biomedical Sciences, School of Medicine, University of California Riverside, Riverside, CA 92521.