Molecular Basis of Drug Recognition by EmrE
Li, J., Sae Her, A., Besch, A., Ramirez, B., Crames, M., Banigan, J.R., Mueller, C., Marsiglia, W.M., Zhang, Y., Traaseth, N.J.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| SMR family multidrug efflux protein EmrE | 110 | Escherichia coli | Mutation(s): 1 Gene Names: emrE Membrane Entity: Yes | 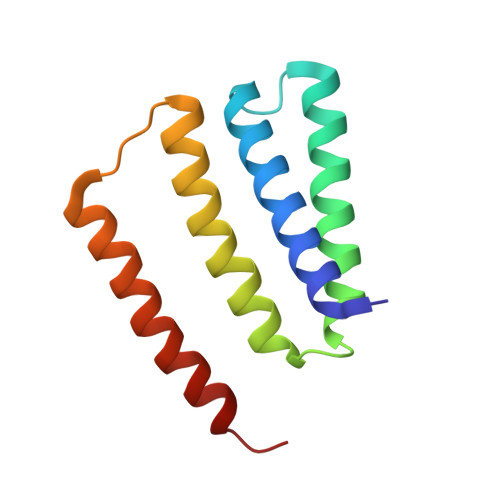 | |
UniProt | |||||
Find proteins for A0A2X7QID6 (Escherichia coli) Explore A0A2X7QID6 Go to UniProtKB: A0A2X7QID6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A2X7QID6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| SMR family multidrug efflux protein EmrE | 110 | Escherichia coli | Mutation(s): 0 Gene Names: emrE Membrane Entity: Yes | 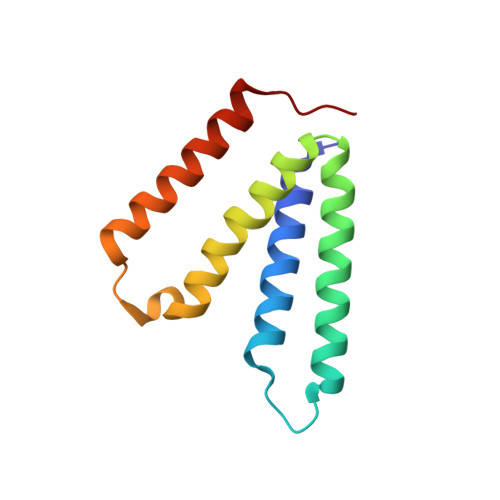 | |
UniProt | |||||
Find proteins for A0A2X7QID6 (Escherichia coli) Explore A0A2X7QID6 Go to UniProtKB: A0A2X7QID6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A2X7QID6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute Of Allergy and Infectious Diseases (NIH/NIAID) | United States | R01AI108889 |