Characterisation of a novel antimalarial agent targeting haemaglobin digestion that shows cross-species reactivity and excellent in vivo properties.
de Koning-Ward, T.F., Drinkwater, N., Edgar, R.C.S., McGowan, S., Scammells, P.J.(2024) mBio
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2024) mBio
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Leucine aminopeptidase | 528 | Plasmodium falciparum | Mutation(s): 4 Gene Names: LAP, PF3D7_1446200 EC: 3.4.11.1 (PDB Primary Data), 3.4.13 (PDB Primary Data) | 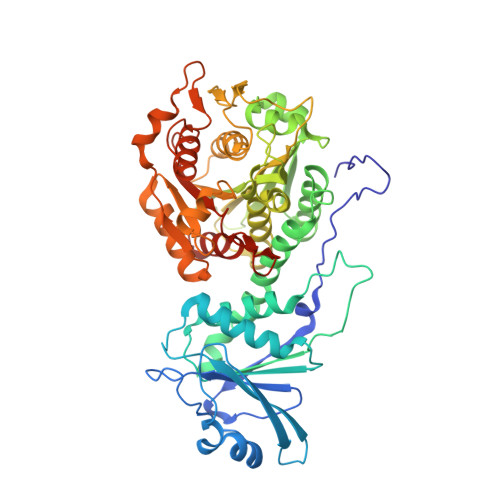 | |
UniProt | |||||
Find proteins for Q8IL11 (Plasmodium falciparum (isolate 3D7)) Explore Q8IL11 Go to UniProtKB: Q8IL11 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8IL11 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 1PE Query on 1PE | CA [auth C] CB [auth G] DA [auth C] DB [auth G] EA [auth C] | PENTAETHYLENE GLYCOL C10 H22 O6 JLFNLZLINWHATN-UHFFFAOYSA-N |  | ||
| SO4 Query on SO4 | AC [auth K] BA [auth C] BB [auth G] BC [auth L] FA [auth C] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| ZN Query on ZN | AB [auth G] CC [auth L] DC [auth L] GA [auth D] GB [auth H] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| CO3 Query on CO3 | AA [auth C] EC [auth L] IA [auth D] IB [auth H] NB [auth I] | CARBONATE ION C O3 BVKZGUZCCUSVTD-UHFFFAOYSA-L |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 174.335 | α = 90 |
| b = 176.725 | β = 90 |
| c = 225.21 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| Aimless | data scaling |
| XDS | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Health and Medical Research Council (NHMRC, Australia) | Australia | 1185354 |