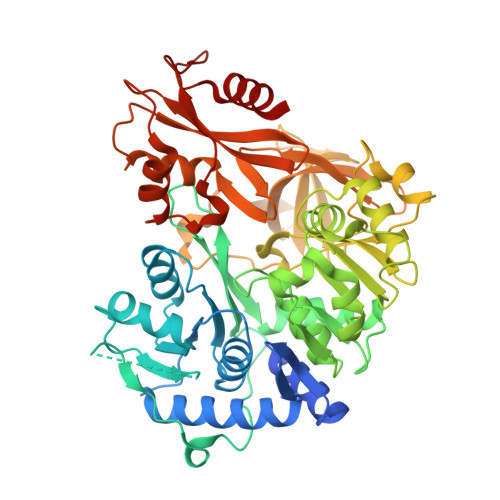
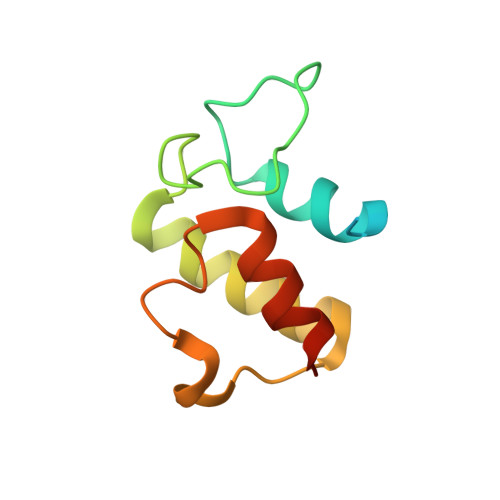
Structural Basis of Amide-Forming Adenylation Enzyme VinM in Vicenistatin Biosynthesis.
Miyanaga, A., Nagata, K., Nakajima, J., Chisuga, T., Kudo, F., Eguchi, T.(2023) ACS Chem Biol 18: 2343-2348
- PubMed: 37870408
- DOI: https://doi.org/10.1021/acschembio.3c00517
- Primary Citation of Related Structures:
8K4R - PubMed Abstract:
Adenylation enzymes activate amino acid substrates to aminoacyl adenylates and generally transfer this moiety onto the thiol group of the phosphopantetheine arm of a carrier protein for the selective incorporation of aminoacyl building blocks in natural product biosynthesis. In contrast to the canonical thioester-forming adenylation enzymes, the amide-forming adenylation enzyme VinM transfers an l-alanyl group onto the amino group of the aminoacyl unit attached to the phosphopantetheine arm of the carrier protein VinL to generate dipeptidyl-VinL in vicenistatin biosynthesis. It is unclear how VinM distinguishes aminoacyl-VinL from VinL for amide bond formation. Herein we describe structural and biochemical analyses of VinM. We determined the crystal structure of VinM in complex with VinL using a designed pantetheine-type cross-linking probe. The VinM-VinL complex structure in combination with site-directed mutagenesis analysis revealed that the interactions with both the phosphopantetheine arm and VinL are critical for the amide-forming activity of VinM.
- Department of Chemistry, Tokyo Institute of Technology, Tokyo 152-8551, Japan.
Organizational Affiliation: