TRIM7 Restricts Coxsackievirus and Norovirus Infection by Detecting the C-Terminal Glutamine Generated by 3C Protease Processing.
Luptak, J., Mallery, D.L., Jahun, A.S., Albecka, A., Clift, D., Ather, O., Slodkowicz, G., Goodfellow, I., James, L.C.(2022) Viruses 14
- PubMed: 35893676
- DOI: https://doi.org/10.3390/v14081610
- Primary Citation of Related Structures:
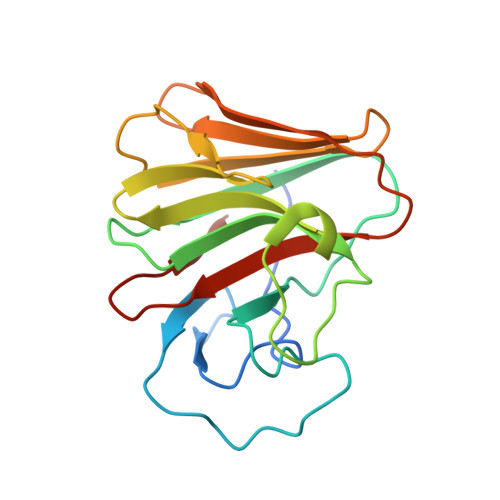
8A5L, 8A5M, 8A8X - PubMed Abstract:
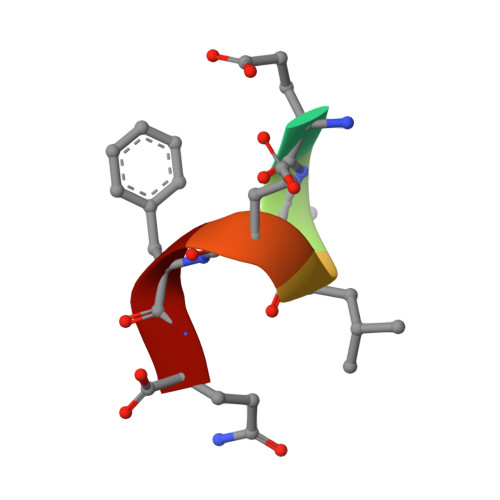
TRIM7 catalyzes the ubiquitination of multiple substrates with unrelated biological functions. This cross-reactivity is at odds with the specificity usually displayed by enzymes, including ubiquitin ligases. Here we show that TRIM7's extreme substrate promiscuity is due to a highly unusual binding mechanism, in which the PRYSPRY domain captures any ligand with a C-terminal helix that terminates in a hydrophobic residue followed by a glutamine. Many of the non-structural proteins found in RNA viruses contain C-terminal glutamines as a result of polyprotein cleavage by 3C protease. This viral processing strategy generates novel substrates for TRIM7 and explains its ability to inhibit Coxsackie virus and norovirus replication. In addition to viral proteins, cellular proteins such as glycogenin have evolved C-termini that make them a TRIM7 substrate. The 'helix-ΦQ' degron motif recognized by TRIM7 is reminiscent of the N-end degron system and is found in ~1% of cellular proteins. These features, together with TRIM7's restricted tissue expression and lack of immune regulation, suggest that viral restriction may not be its physiological function.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge CB2 0QH, UK.