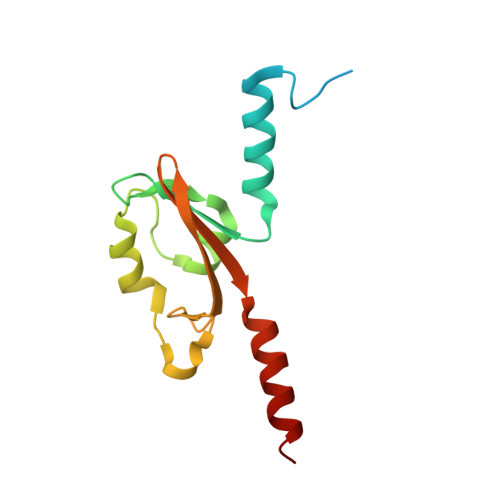
PAS domain of flagellar histidine kinase FlrB has a unique architecture and binds heme as a sensory ligand in an unconventional fashion.
Mukherjee, P., Agarwal, S., Mallick, S.B., Dasgupta, J.(2024) Structure 32: 200-216.e5
- PubMed: 38157857
- DOI: https://doi.org/10.1016/j.str.2023.11.014
- Primary Citation of Related Structures:
7YRT - PubMed Abstract:
Phosphorylation of the σ 54 -dependent transcription activator FlrC by the sensor histidine kinase FlrB is essential for flagellar synthesis of Vibrio cholerae. Despite that, the structure, sensory signal, and mechanistic basis of function of FlrB were elusive. Here, we report the crystal structure of the sensory PAS domain of FlrB in its functional dimeric state that exhibits a unique architecture. Series of biochemical/biophysical experiments on different constructs and mutants established that heme binds hydrophobically as sensory ligand in the shallow ligand-binding cleft of FlrB-PAS without axial coordination. Intriguingly, ATP binding to the C-terminal ATP-binding (CA) domain assists PAS domain to bind heme, vis-à-vis, heme binding to the PAS facilitates ATP binding to the CA domain. We hypothesize that synergistic binding of heme and ATP triggers conformational signaling in FlrB, leading to downstream flagellar gene transcription. Enhanced swimming motility of V. cholerae with increased heme uptake supports this proposition.
Organizational Affiliation:
Department of Biotechnology, St. Xavier's College (Autonomous), 30 Mother Teresa Sarani, Kolkata 700016, India.