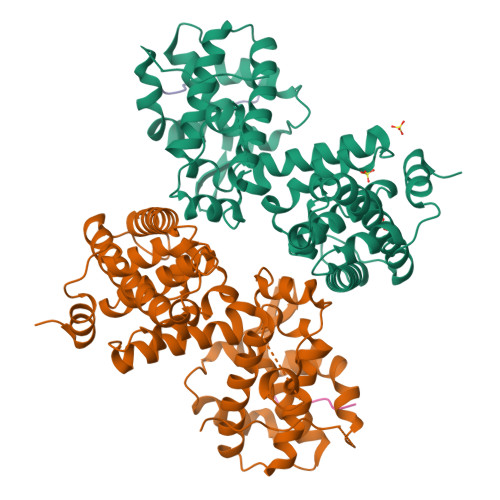
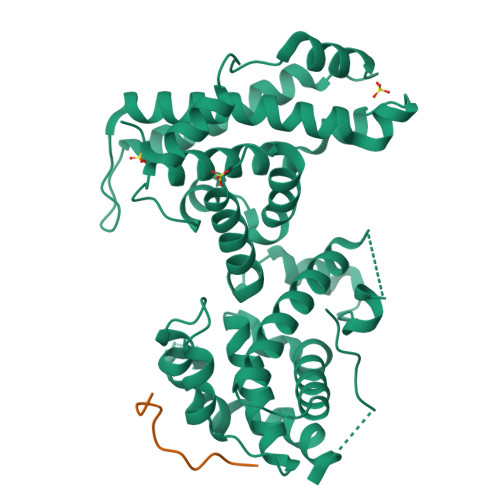
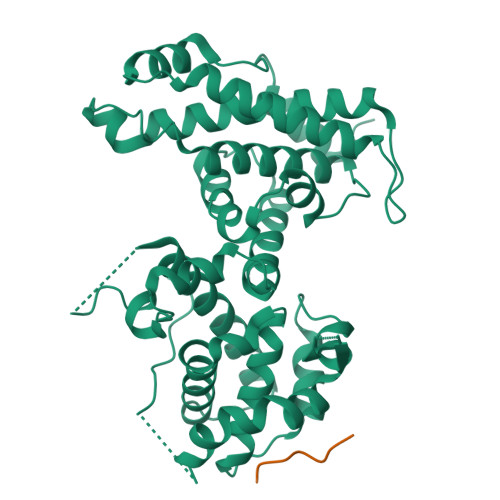
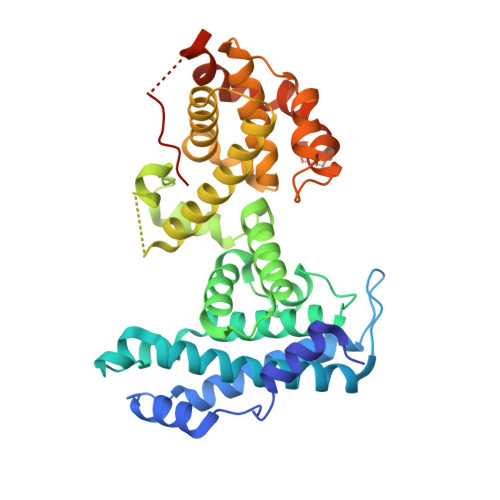
Structural basis for tunable affinity and specificity of LxCxE-dependent protein interactions with the retinoblastoma protein family.
Putta, S., Alvarez, L., Ludtke, S., Sehr, P., Muller, G.A., Fernandez, S.M., Tripathi, S., Lewis, J., Gibson, T.J., Chemes, L.B., Rubin, S.M.(2022) Structure 30: 1340
- PubMed: 35716663
- DOI: https://doi.org/10.1016/j.str.2022.05.019
- Primary Citation of Related Structures:
7SMC, 7SMD, 7SME, 7SMF - PubMed Abstract:
The retinoblastoma protein (Rb) and its homologs p107 and p130 are critical regulators of gene expression during the cell cycle and are commonly inactivated in cancer. Rb proteins use their "pocket domain" to bind an LxCxE sequence motif in other proteins, many of which function with Rb proteins to co-regulate transcription. Here, we present binding data and crystal structures of the p107 pocket domain in complex with LxCxE peptides from the transcriptional co-repressor proteins HDAC1, ARID4A, and EID1. Our results explain why Rb and p107 have weaker affinity for cellular LxCxE proteins compared with the E7 protein from human papillomavirus, which has been used as the primary model for understanding LxCxE motif interactions. Our structural and mutagenesis data also identify and explain differences in Rb and p107 affinities for some LxCxE-containing sequences. Our study provides new insights into how Rb proteins bind their cell partners with varying affinity and specificity.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California, Santa Cruz, CA 95064, USA.