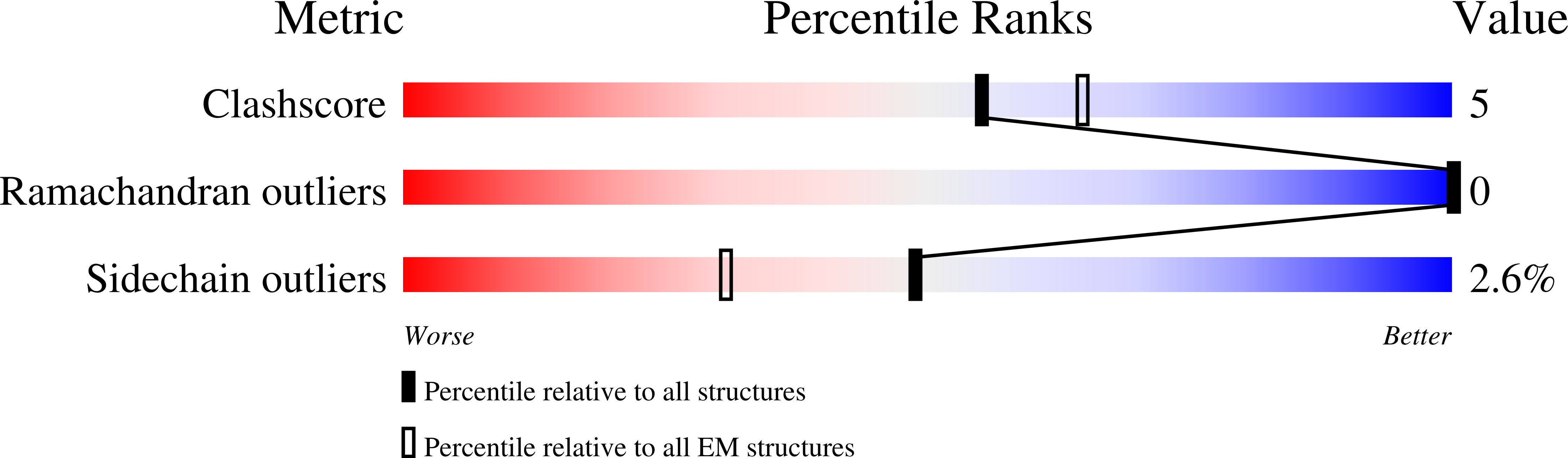The structural basis of Rubisco phase separation in the pyrenoid.
He, S., Chou, H.T., Matthies, D., Wunder, T., Meyer, M.T., Atkinson, N., Martinez-Sanchez, A., Jeffrey, P.D., Port, S.A., Patena, W., He, G., Chen, V.K., Hughson, F.M., McCormick, A.J., Mueller-Cajar, O., Engel, B.D., Yu, Z., Jonikas, M.C.(2020) Nat Plants 6: 1480-1490
- PubMed: 33230314
- DOI: https://doi.org/10.1038/s41477-020-00811-y
- Primary Citation of Related Structures:
7JFO, 7JN4, 7JSX - PubMed Abstract:
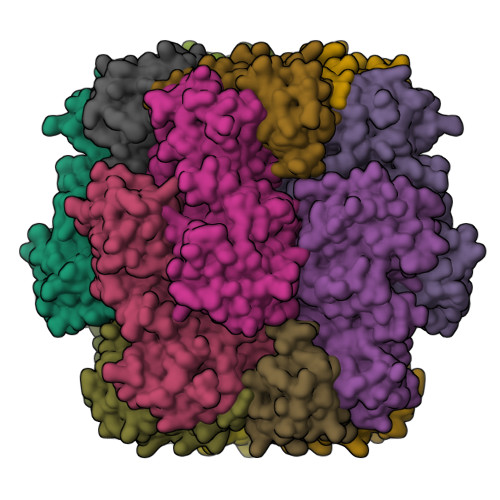
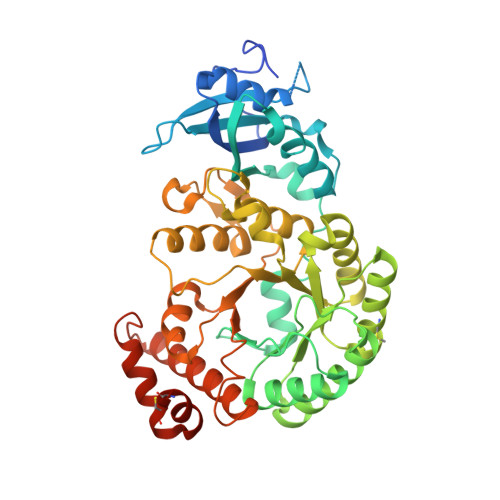
Approximately one-third of global CO 2 fixation occurs in a phase-separated algal organelle called the pyrenoid. The existing data suggest that the pyrenoid forms by the phase separation of the CO 2 -fixing enzyme Rubisco with a linker protein; however, the molecular interactions underlying this phase separation remain unknown. Here we present the structural basis of the interactions between Rubisco and its intrinsically disordered linker protein Essential Pyrenoid Component 1 (EPYC1) in the model alga Chlamydomonas reinhardtii. We find that EPYC1 consists of five evenly spaced Rubisco-binding regions that share sequence similarity. Single-particle cryo-electron microscopy of these regions in complex with Rubisco indicates that each Rubisco holoenzyme has eight binding sites for EPYC1, one on each Rubisco small subunit. Interface mutations disrupt binding, phase separation and pyrenoid formation. Cryo-electron tomography supports a model in which EPYC1 and Rubisco form a codependent multivalent network of specific low-affinity bonds, giving the matrix liquid-like properties. Our results advance the structural and functional understanding of the phase separation underlying the pyrenoid, an organelle that plays a fundamental role in the global carbon cycle.
Organizational Affiliation:
Department of Molecular Biology, Princeton University, Princeton, NJ, USA.