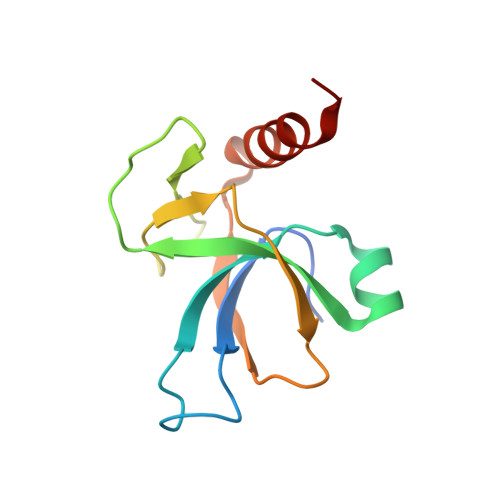
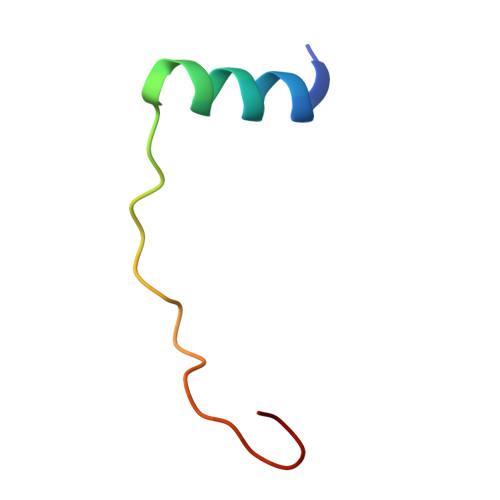
Role of PemI in the Staphylococcus aureus PemIK toxin-antitoxin complex: PemI controls PemK by acting as a PemK loop mimic.
Kim, D.H., Kang, S.M., Baek, S.M., Yoon, H.J., Jang, D.M., Kim, H.S., Lee, S.J., Lee, B.J.(2022) Nucleic Acids Res 50: 2319-2333
- PubMed: 35141752
- DOI: https://doi.org/10.1093/nar/gkab1288
- Primary Citation of Related Structures:
7EWI, 7EWJ - PubMed Abstract:
Staphylococcus aureus is a notorious and globally distributed pathogenic bacterium. New strategies to develop novel antibiotics based on intrinsic bacterial toxin-antitoxin (TA) systems have been recently reported. Because TA systems are present only in bacteria and not in humans, these distinctive systems are attractive targets for developing antibiotics with new modes of action. S. aureus PemIK is a type II TA system, comprising the toxin protein PemK and the labile antitoxin protein PemI. Here, we determined the crystal structures of both PemK and the PemIK complex, in which PemK is neutralized by PemI. Our biochemical approaches, including fluorescence quenching and polarization assays, identified Glu20, Arg25, Thr48, Thr49, and Arg84 of PemK as being important for RNase function. Our study indicates that the active site and RNA-binding residues of PemK are covered by PemI, leading to unique conformational changes in PemK accompanied by repositioning of the loop between β1 and β2. These changes can interfere with RNA binding by PemK. Overall, PemK adopts particular open and closed forms for precise neutralization by PemI. This structural and functional information on PemIK will contribute to the discovery and development of novel antibiotics in the form of peptides or small molecules inhibiting direct binding between PemI and PemK.
Organizational Affiliation:
Jeju Research Institute of Pharmaceutical Sciences, College of Pharmacy, Jeju National University, Jeju 63243, Republic of Korea.