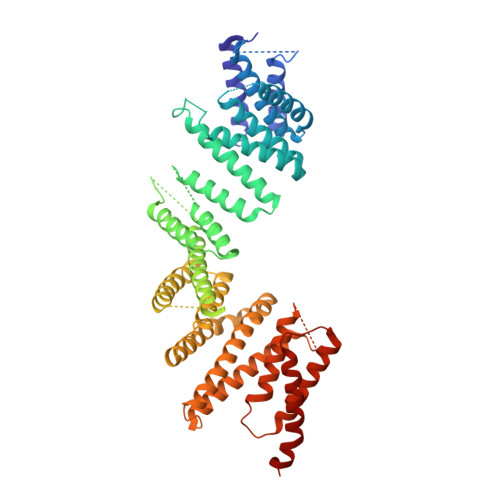
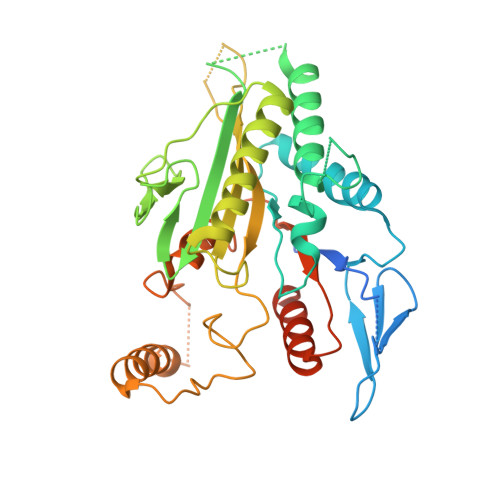
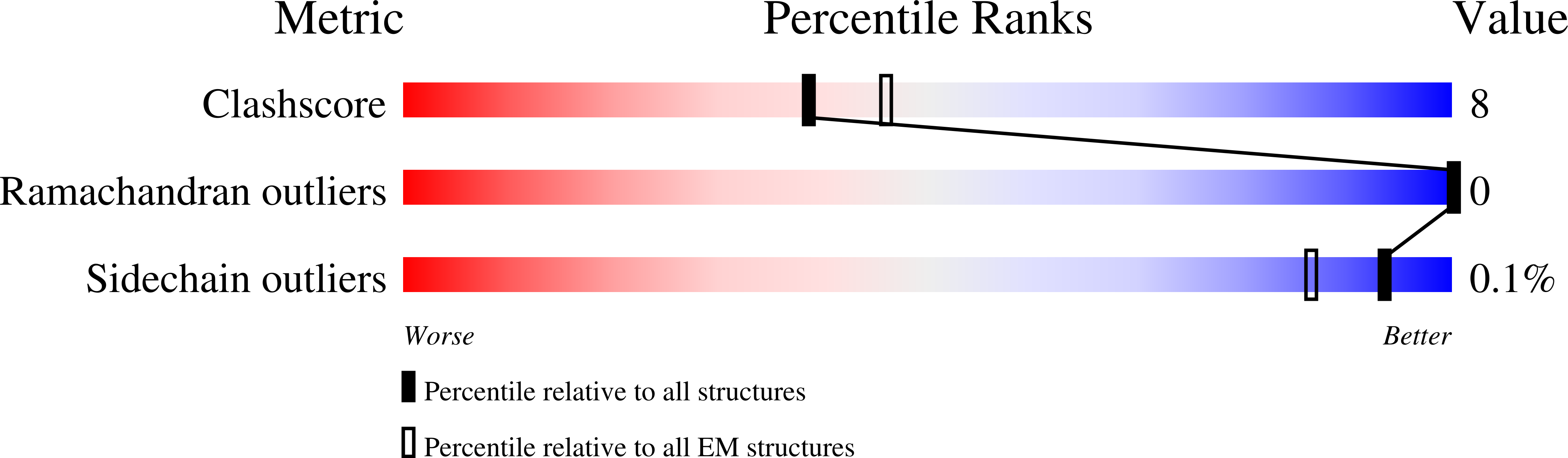The mechanism of kinesin inhibition by kinesin-binding protein.
Atherton, J., Hummel, J.J., Olieric, N., Locke, J., Pena, A., Rosenfeld, S.S., Steinmetz, M.O., Hoogenraad, C.C., Moores, C.A.(2020) Elife 9
- PubMed: 33252036
- DOI: https://doi.org/10.7554/eLife.61481
- Primary Citation of Related Structures:
6ZPG, 6ZPH, 6ZPI - PubMed Abstract:
Subcellular compartmentalisation is necessary for eukaryotic cell function. Spatial and temporal regulation of kinesin activity is essential for building these local environments via control of intracellular cargo distribution. Kinesin-binding protein (KBP) interacts with a subset of kinesins via their motor domains, inhibits their microtubule (MT) attachment, and blocks their cellular function. However, its mechanisms of inhibition and selectivity have been unclear. Here we use cryo-electron microscopy to reveal the structure of KBP and of a KBP-kinesin motor domain complex. KBP is a tetratricopeptide repeat-containing, right-handed α-solenoid that sequesters the kinesin motor domain's tubulin-binding surface, structurally distorting the motor domain and sterically blocking its MT attachment. KBP uses its α-solenoid concave face and edge loops to bind the kinesin motor domain, and selected structure-guided mutations disrupt KBP inhibition of kinesin transport in cells. The KBP-interacting motor domain surface contains motifs exclusively conserved in KBP-interacting kinesins, suggesting a basis for kinesin selectivity.
Organizational Affiliation:
Randall Centre for Cell and Molecular Biophysics, King's College, London, United Kingdom.