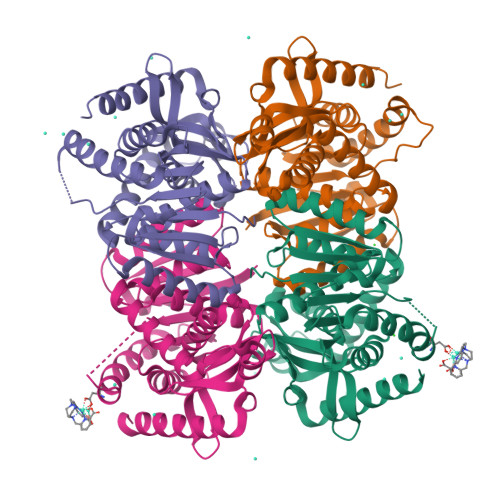
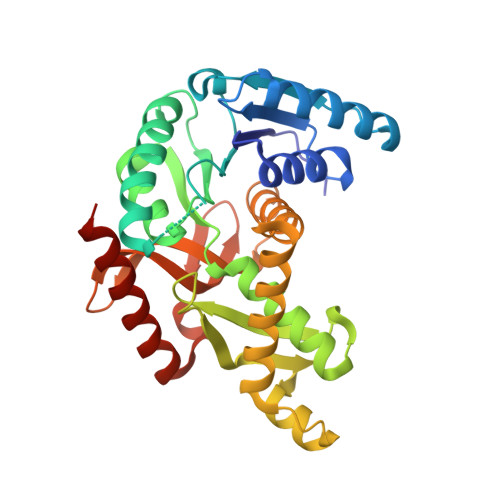
The archaeal LDH-like malate dehydrogenase from Ignicoccus islandicus displays dual substrate recognition, hidden allostery and a non-canonical tetrameric oligomeric organization.
Roche, J., Girard, E., Mas, C., Madern, D.(2019) J Struct Biol 208: 7-17
- PubMed: 31301348
- DOI: https://doi.org/10.1016/j.jsb.2019.07.006
- Primary Citation of Related Structures:
6QSS - PubMed Abstract:
The NAD(P)-dependent malate dehydrogenases (MalDHs) and NAD-dependent lactate dehydrogenases (LDHs) are homologous enzymes involved in central metabolism. They display a common protein fold and the same catalytic mechanism, yet have a stringent capacity to discriminate between their respective substrates. The MalDH/LDH superfamily is divided into several phylogenetically related groups. It has been shown that the canonical LDHs and LDH-like group of MalDHs are primarily tetrameric enzymes that diverged from a common ancestor. In order to gain understanding of the evolutionary history of the LDHs and MalDHs, the biochemical properties and crystallographic structure of the LDH-like MalDH from the hyperthermophilic archaeon Ignicoccus islandicus (I. isl) were determined. I. isl MalDH recognizes oxaloacetate as main substrate, but it is also able to use pyruvate. Surprisingly, with pyruvate, the enzymatic activity profile looks like that of allosteric LDHs, suggesting a hidden allosteric capacity in a MalDH. The I. isl MalDH tetrameric structure in the apo state is considerably different from those of canonical LDH-like MalDHs and LDHs, representing an alternative oligomeric organization. A comparison with MalDH and LDH counterparts provides strong evidence that the divergence between allosteric and non-allosteric members of the superfamily involves homologs with intermediate, atypical properties.
Organizational Affiliation:
Univ. Grenoble Alpes, CEA, CNRS, IBS, 38000 Grenoble, France.