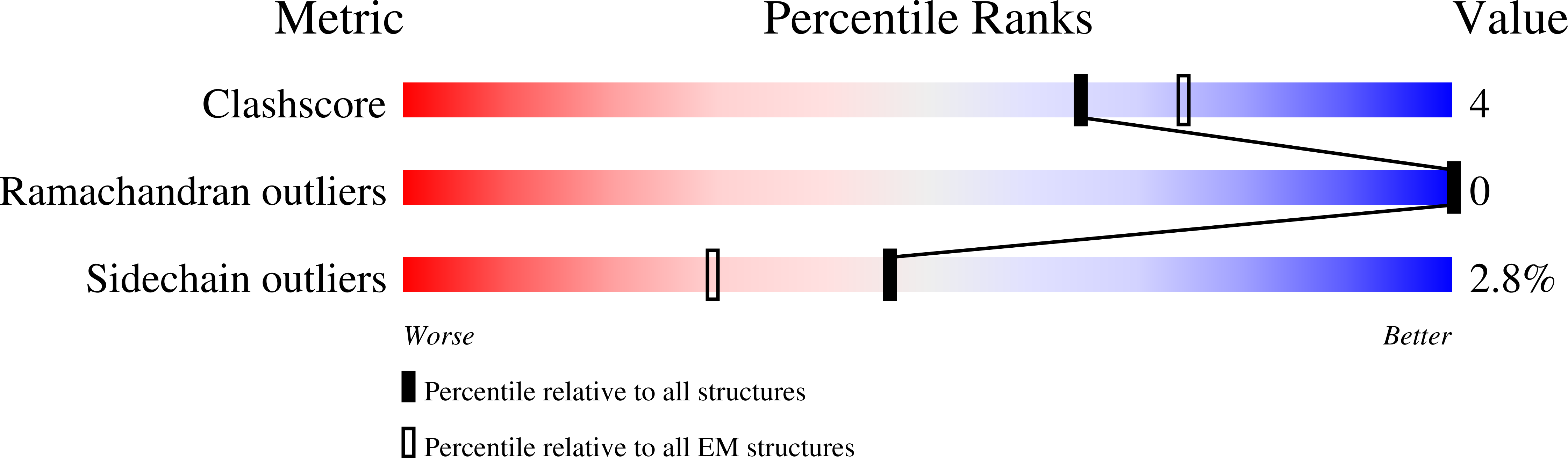Structural Basis for Tetherin Antagonism as a Barrier to Zoonotic Lentiviral Transmission.
Buffalo, C.Z., Sturzel, C.M., Heusinger, E., Kmiec, D., Kirchhoff, F., Hurley, J.H., Ren, X.(2019) Cell Host Microbe 26: 359-368.e8
- PubMed: 31447307
- DOI: https://doi.org/10.1016/j.chom.2019.08.002
- Primary Citation of Related Structures:
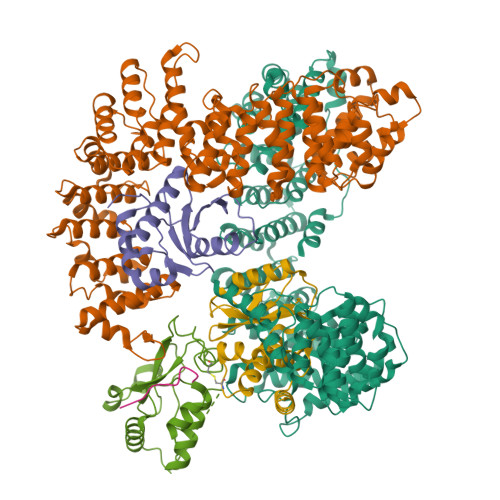
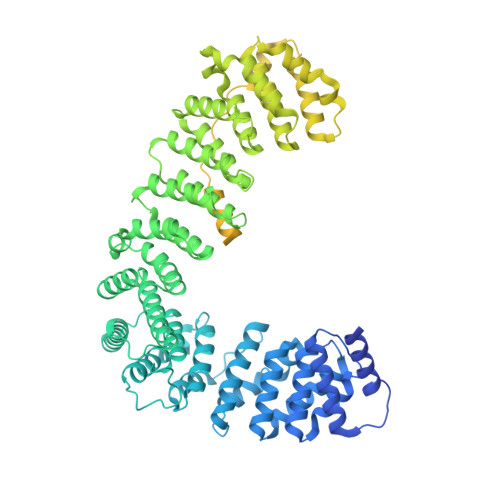
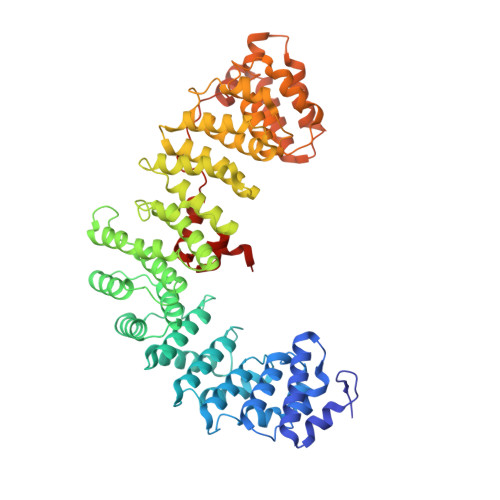
6OWT - PubMed Abstract:
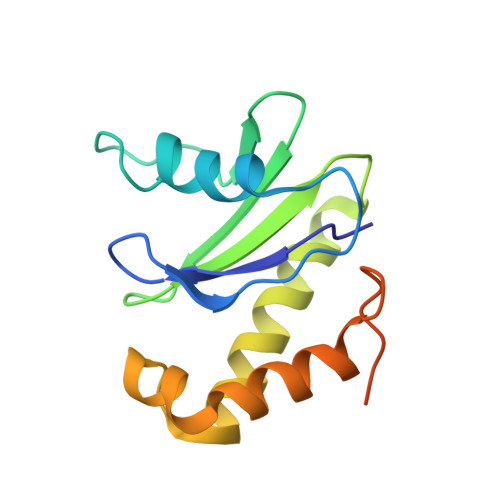
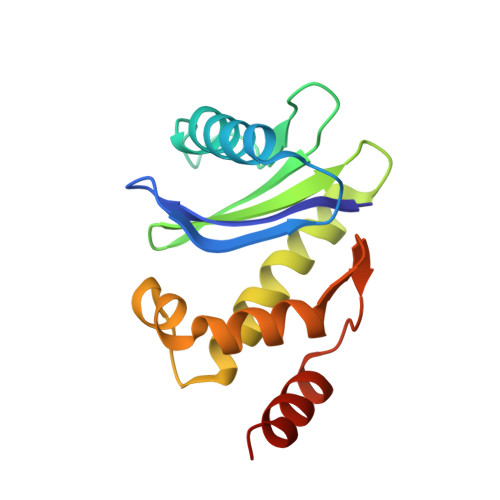
Tetherin is a host defense factor that physically prevents virion release from the plasma membrane. The Nef accessory protein of simian immunodeficiency virus (SIV) engages the clathrin adaptor AP-2 to downregulate tetherin via its DIWK motif. As human tetherin lacks DIWK, antagonism of tetherin by Nef is a barrier to simian-human transmission of non-human primate lentiviruses. To determine the molecular basis for tetherin counteraction, we reconstituted the AP-2 complex with a simian tetherin and SIV Nef and determined its structure by cryoelectron microscopy (cryo-EM). Nef refolds the first α-helix of the β2 subunit of AP-2 to a β hairpin, creating a binding site for the DIWK sequence. The tetherin binding site in Nef is distinct from those of most other Nef substrates, including MHC class I, CD3, and CD4 but overlaps with the site for the restriction factor SERINC5. This structure explains the dependence of SIVs on tetherin DIWK and consequent barrier to human transmission.
Organizational Affiliation:
Department of Molecular and Cell Biology and California Institute for Quantitative Biosciences, University of California, Berkeley, Berkeley, CA 94720, USA.