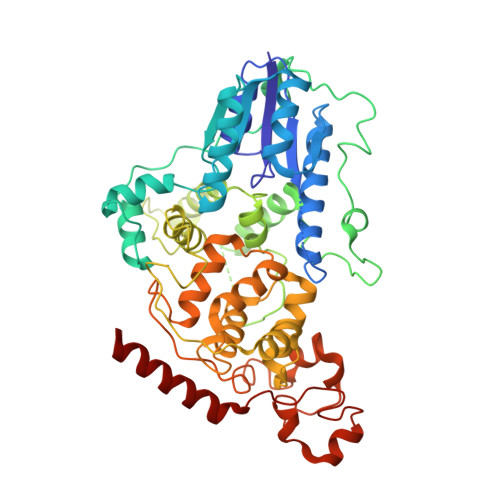
Dynamics at the serine loop underlie differential affinity of cryptochromes for CLOCK:BMAL1 to control circadian timing.
Fribourgh, J.L., Srivastava, A., Sandate, C.R., Michael, A.K., Hsu, P.L., Rakers, C., Nguyen, L.T., Torgrimson, M.R., Parico, G.C.G., Tripathi, S., Zheng, N., Lander, G.C., Hirota, T., Tama, F., Partch, C.L.(2020) Elife 9
- PubMed: 32101164
- DOI: https://doi.org/10.7554/eLife.55275
- Primary Citation of Related Structures:
6OF7 - PubMed Abstract:
Mammalian circadian rhythms are generated by a transcription-based feedback loop in which CLOCK:BMAL1 drives transcription of its repressors (PER1/2, CRY1/2), which ultimately interact with CLOCK:BMAL1 to close the feedback loop with ~24 hr periodicity. Here we pinpoint a key difference between CRY1 and CRY2 that underlies their differential strengths as transcriptional repressors. Both cryptochromes bind the BMAL1 transactivation domain similarly to sequester it from coactivators and repress CLOCK:BMAL1 activity. However, we find that CRY1 is recruited with much higher affinity to the PAS domain core of CLOCK:BMAL1, allowing it to serve as a stronger repressor that lengthens circadian period. We discovered a dynamic serine-rich loop adjacent to the secondary pocket in the photolyase homology region (PHR) domain that regulates differential binding of cryptochromes to the PAS domain core of CLOCK:BMAL1. Notably, binding of the co-repressor PER2 remodels the serine loop of CRY2, making it more CRY1-like and enhancing its affinity for CLOCK:BMAL1.
- Department of Chemistry and Biochemistry, University of California Santa Cruz, Santa Cruz, United States.
Organizational Affiliation: