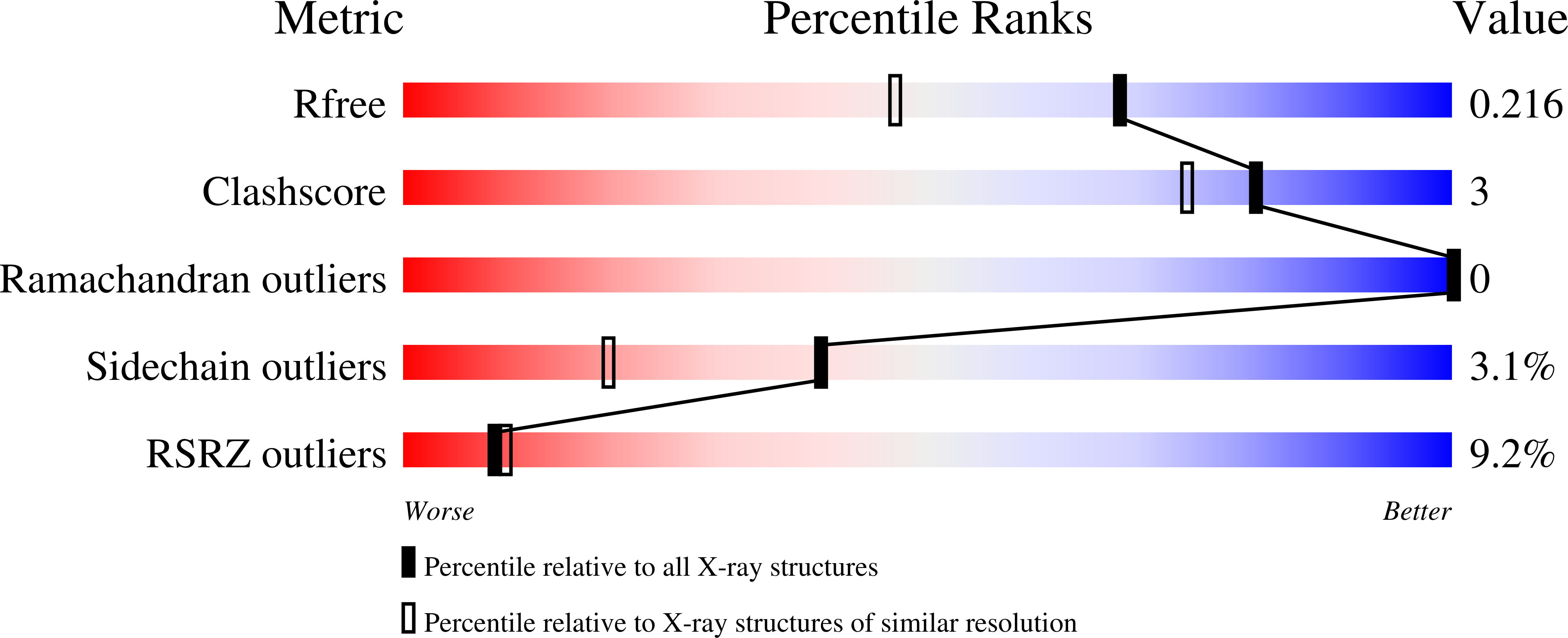
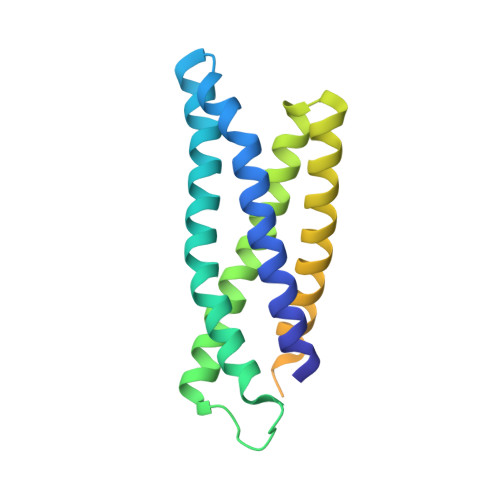
Large protein organelles form a new iron sequestration system with high storage capacity.
Giessen, T.W., Orlando, B.J., Verdegaal, A.A., Chambers, M.G., Gardener, J., Bell, D.C., Birrane, G., Liao, M., Silver, P.A.(2019) Elife 8
- PubMed: 31282860
- DOI: https://doi.org/10.7554/eLife.46070
- Primary Citation of Related Structures:
6N63, 6NJ8 - PubMed Abstract:
Iron storage proteins are essential for cellular iron homeostasis and redox balance. Ferritin proteins are the major storage units for bioavailable forms of iron. Some organisms lack ferritins, and it is not known how they store iron. Encapsulins, a class of protein-based organelles, have recently been implicated in microbial iron and redox metabolism. Here, we report the structural and mechanistic characterization of a 42 nm two-component encapsulin-based iron storage compartment from Quasibacillus thermotolerans . Using cryo-electron microscopy and x-ray crystallography, we reveal the assembly principles of a thermostable T = 4 shell topology and its catalytic ferroxidase cargo and show interactions underlying cargo-shell co-assembly. This compartment has an exceptionally large iron storage capacity storing over 23,000 iron atoms. Our results reveal a new approach for survival in diverse habitats with limited or fluctuating iron availability via an iron storage system able to store 10 to 20 times more iron than ferritin.
Organizational Affiliation:
Department of Systems Biology, Harvard Medical School, Boston, United States.