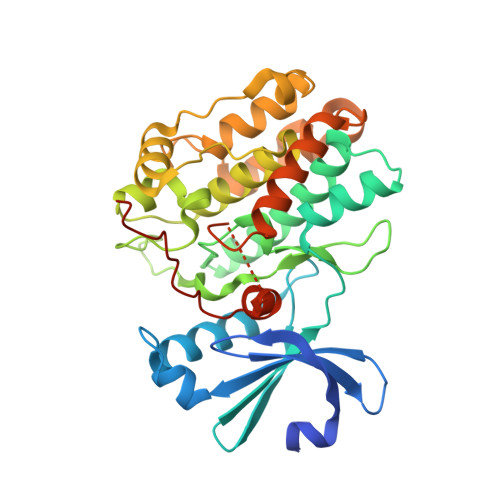
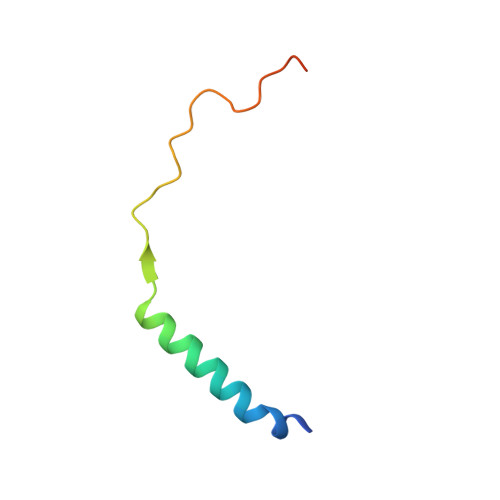
Structural Basis for the High-Affinity Interaction between CASK and Mint1.
Wu, X., Cai, Q., Chen, Y., Zhu, S., Mi, J., Wang, J., Zhang, M.(2020) Structure 28: 664-673.e3
- PubMed: 32348748
- DOI: https://doi.org/10.1016/j.str.2020.04.001
- Primary Citation of Related Structures:
6LNM - PubMed Abstract:
CASK forms an evolutionarily conserved tripartite complex with Mint1 and Veli critical for neuronal synaptic transmission and cell polarity. The CASK CaM kinase (CaMK) domain, in addition to interacting with Mint1, can also bind to many different target proteins, although the mechanism governing CASK-CaMK/target interaction selectivity is unclear. Here, we demonstrate that an extended sequence in the N-terminal unstructured region of Mint1 binds to CASK-CaMK with a dissociation constant of ∼7.5 nM. The high-resolution crystal structure of CASK-CaMK in complex with this Mint1 fragment reveals that the C-lobe of CASK-CaMK binds to a short sequence common to known CaMK targets and the N-lobe of CaMK engages an α helix that is unique to Mint1. Biochemical experiments together with structural analysis reveal that the CASK and Mint1 interaction is not regulated by Ca 2+ /CaM. The CASK/Mint1 complex structure provides mechanistic explanations for several CASK mutations identified in patients with brain disorders and cancers.
Organizational Affiliation:
Division of Life Science, State Key Laboratory of Molecular Neuroscience, Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong, China.