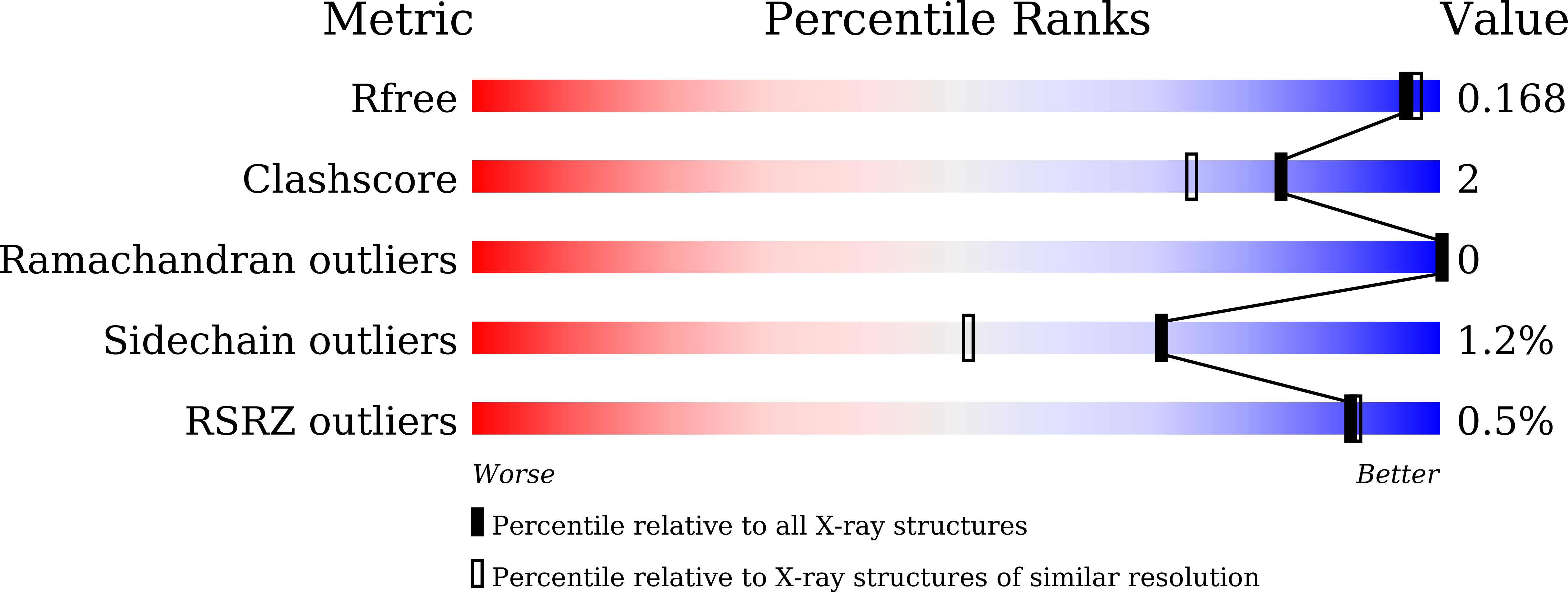
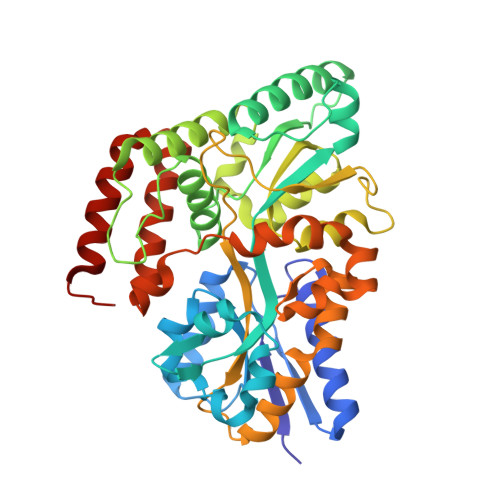
Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Chandravanshi, M., Gogoi, P., Kanaujia, S.P.(2020) FEBS J 287: 1576-1597
- PubMed: 31608555
- DOI: https://doi.org/10.1111/febs.15093
- Primary Citation of Related Structures:
6J9W, 6J9Y, 6JAD, 6JAG, 6JAH, 6JAI, 6JAL, 6JAM, 6JAN, 6JAO, 6JAP, 6JAQ, 6JAR, 6JAZ, 6JB0, 6JB4, 6JBA, 6JBB, 6JBE - PubMed Abstract:
Carbohydrate (or sugar) molecules are extremely diverse regarding their length, linkage and epimeric state. Selective acquisition of these molecules inside the cell is achieved by the substrate (or solute)-binding protein of ATP-binding cassette (ABC) transport system. However, the molecular mechanism underlying the selective transport of diverse carbohydrates remains unclear mainly owing to their structural complexity and stereochemistry. This study reports crystal structures of an α-glycoside-binding protein (αGlyBP, ORF ID: TTHA0356 from Thermus thermophilus HB8) in complex with disaccharide α-glycosides namely trehalose (α-1,1), sucrose (α-1,2), maltose (α-1,4), palatinose (α-1,6) and glucose within a resolution range of 1.6-2.0 Å. Despite transporting multiple types of sugars, αGlyBP maintains its stereoselectivity for both glycosidic linkage as well as an epimeric hydroxyl group. Out of the two subsites identified in the active-site pocket, subsite B which accommodates the glucose and glycosyl unit of disaccharide α-glycosides is highly conserved. In addition, structural data confirms the paradoxical behavior of glucose, where it replaces the high-affinity ligand(s) (disaccharide α-glycosides) from the active site of the protein. Comparative assessment of open and closed conformations of αGlyBP along with mutagenic and thermodynamic studies identifies the hinge region as the first interaction site for the ligands. On the other hand, encapsulation of ligand inside the active site is achieved through the N-terminal domain (NTD) movement, whereas the C-terminal domain (CTD) of αGlyBP is identified to be rigid and postulated to be responsible for maintaining the interaction with the transmembrane domain (TMD) during substrate translocation. DATABASE: Structural data are available in RCSB Protein Data Bank under the accession number(s) 6J9W, 6J9Y, 6JAD, 6JAG, 6JAH, 6JAI, 6JAL, 6JAM, 6JAN, 6JAO, 6JAP, 6JAQ, 6JAR, 6JAZ, 6JB0, 6JB4, 6JBA, 6JBB and 6JBE.
Organizational Affiliation:
Department of Biosciences and Bioengineering, Indian Institute of Technology Guwahati, India.