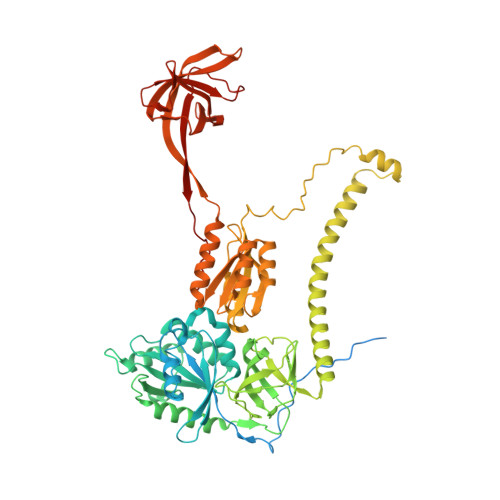
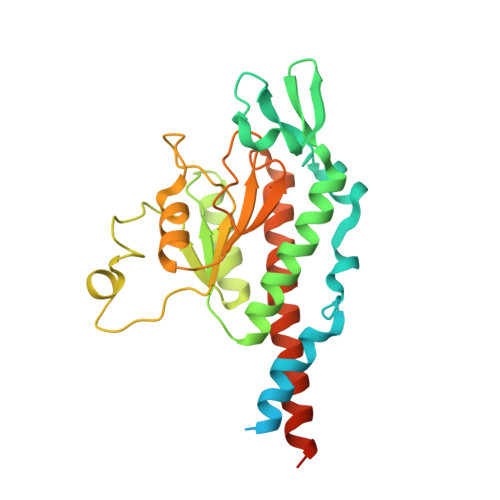
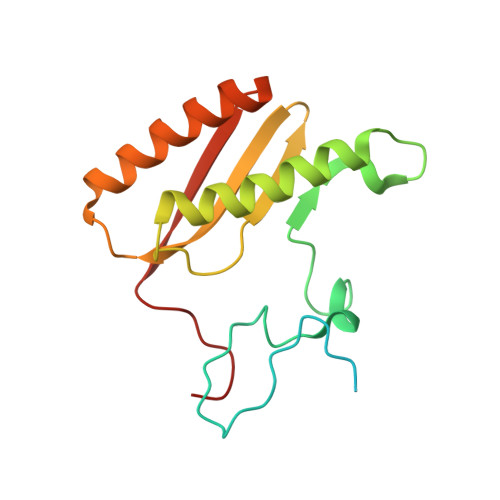
Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM.
Kummer, E., Leibundgut, M., Rackham, O., Lee, R.G., Boehringer, D., Filipovska, A., Ban, N.(2018) Nature 560: 263-267
- PubMed: 30089917
- DOI: https://doi.org/10.1038/s41586-018-0373-y
- Primary Citation of Related Structures:
6GAW, 6GAZ, 6GB2 - PubMed Abstract:
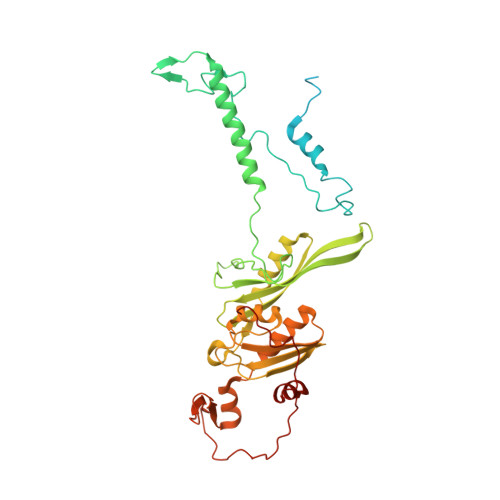
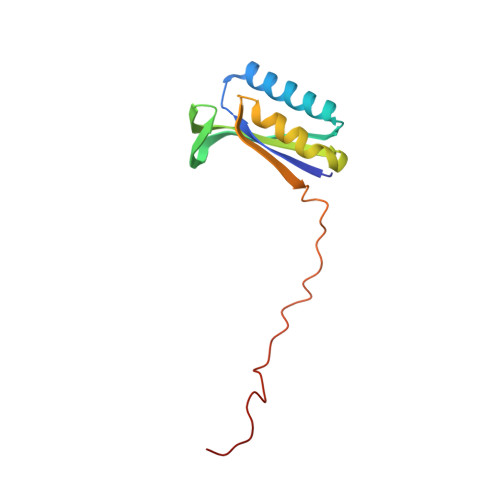
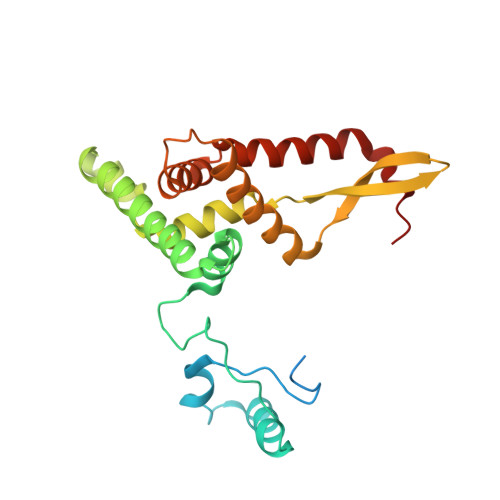
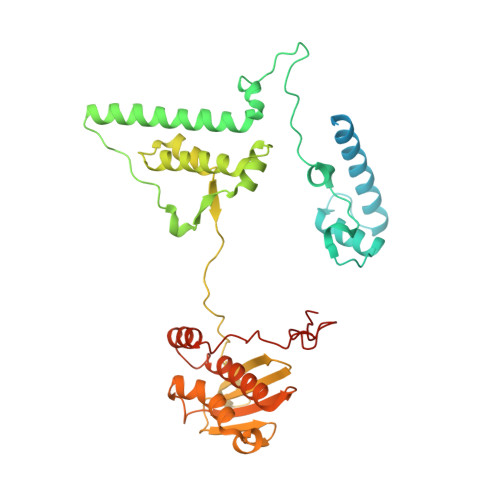
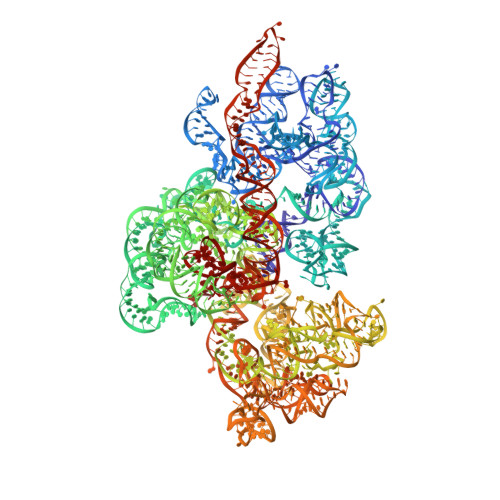
Mitochondria maintain their own specialized protein synthesis machinery, which in mammals is used exclusively for the synthesis of the membrane proteins responsible for oxidative phosphorylation 1,2 . The initiation of protein synthesis in mitochondria differs substantially from bacterial or cytosolic translation systems. Mitochondrial translation initiation lacks initiation factor 1, which is essential in all other translation systems from bacteria to mammals 3,4 . Furthermore, only one type of methionyl transfer RNA (tRNA Met ) is used for both initiation and elongation 4,5 , necessitating that the initiation factor specifically recognizes the formylated version of tRNA Met (fMet-tRNA Met ). Lastly, most mitochondrial mRNAs do not possess 5' leader sequences to promote mRNA binding to the ribosome 2 . There is currently little mechanistic insight into mammalian mitochondrial translation initiation, and it is not clear how mRNA engagement, initiator-tRNA recruitment and start-codon selection occur. Here we determine the cryo-EM structure of the complete translation initiation complex from mammalian mitochondria at 3.2 Å. We describe the function of an additional domain insertion that is present in the mammalian mitochondrial initiation factor 2 (mtIF2). By closing the decoding centre, this insertion stabilizes the binding of leaderless mRNAs and induces conformational changes in the rRNA nucleotides involved in decoding. We identify unique features of mtIF2 that are required for specific recognition of fMet-tRNA Met and regulation of its GTPase activity. Finally, we observe that the ribosomal tunnel in the initiating ribosome is blocked by insertion of the N-terminal portion of mitochondrial protein mL45, which becomes exposed as the ribosome switches to elongation mode and may have an additional role in targeting of mitochondrial ribosomes to the protein-conducting pore in the inner mitochondrial membrane.
Organizational Affiliation:
Department of Biology, Institute of Molecular Biology and Biophysics, ETH Zurich, Zurich, Switzerland.