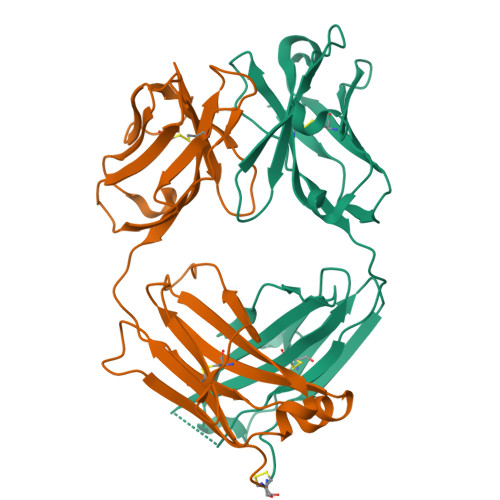
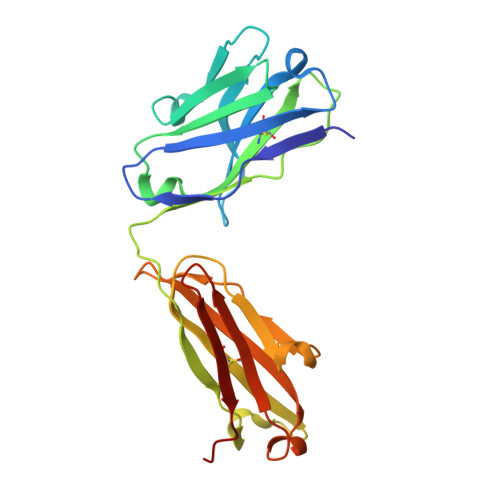
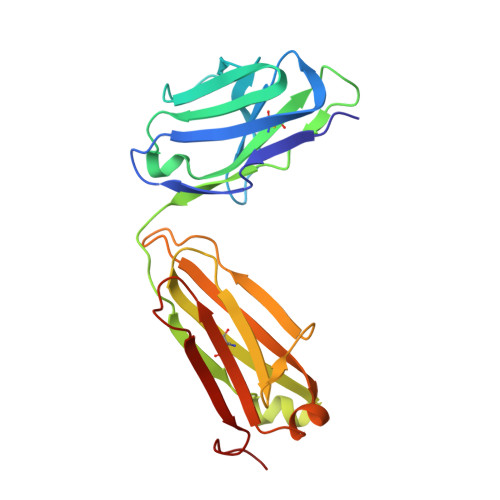
Structural insights into antigen recognition of an anti-beta-(1,6)-beta-(1,3)-D-glucan antibody.
Sung, K.H., Josewski, J., Dubel, S., Blankenfeldt, W., Rau, U.(2018) Sci Rep 8: 13652-13652
- PubMed: 30209318
- DOI: https://doi.org/10.1038/s41598-018-31961-x
- Primary Citation of Related Structures:
6EV1, 6EV2 - PubMed Abstract:
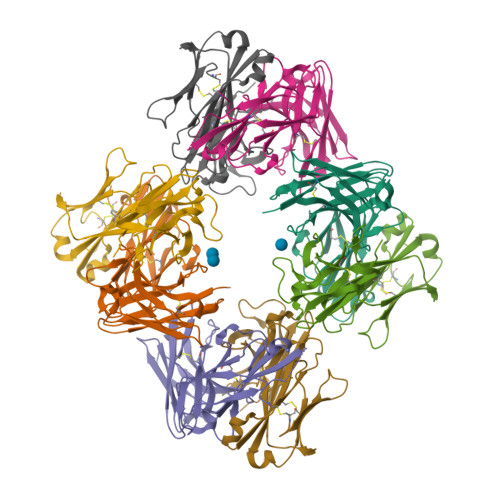
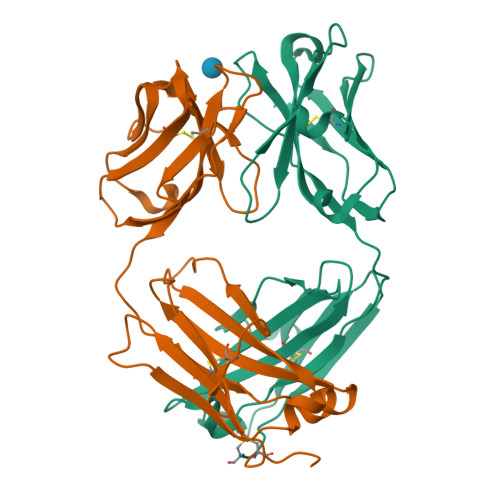
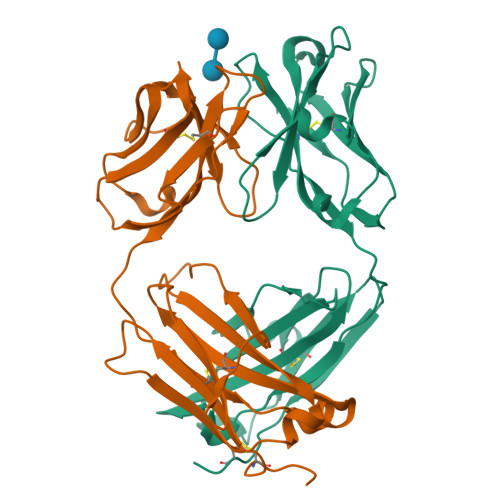
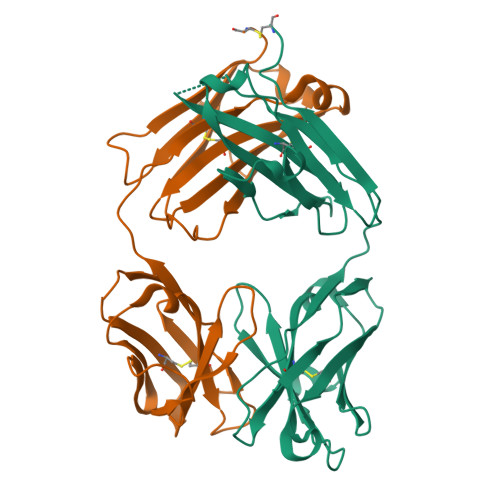
Schizophyllan (SCH) is a high molecular weight homopolysaccharide composed of a β-(1,3)-D-glucan main chain with branching β-(1,6)-bound D-glucose residues. It forms triple helices that are highly stable towards heat and extreme pH, which provides SCH with interesting properties for industrial and medical applications. The recombinant anti-SCH antibody JoJ48C11 recognizes SCH and related β-(1,6)-branched β-(1,3)-D-glucans, but details governing its specificity are not known. Here, we fill this gap by determining crystal structures of the antigen binding fragment (Fab) of JoJ48C11 in the apo form and in complex with the unbranched β-(1,3)-D-glucose hexamer laminarihexaose 3.0 and 2.4 Å resolution, respectively. Together with docking studies, this allowed construction of a JoJ48C11/triple-helical SCH complex, leading to the identification of eight amino acid residues of JoJ48C11 (Tyr27 H , His35 H , Trp47 H , Trp100 H , Asp105 H ; Asp49 L , Lys52 L , Trp90 L ) that contribute to the recognition of glucose units from all three chains of the SCH triple helix. The importance of these amino acids was confirmed by mutagenesis and ELISA-based analysis. Our work provides an explanation for the specific recognition of triple-helical β-(1,6)-branched β-(1,3)-D-glucans by JoJ48C11 and provides another structure example for anti-carbohydrate antibodies.
Organizational Affiliation:
Structure and Function of Proteins, Helmholtz Centre for Infection Research, Inhoffenstraße 7, 38124, Braunschweig, Germany.